Michael Zimmermann
Group Leader
ORCID: 0000-0002-5797-3589
EditMetabolic microbiome–host interactions

Group Leader
ORCID: 0000-0002-5797-3589
EditThe gut microbiota play an essential role in host health, helping to catabolise otherwise indigestible nutrients, engaging in host metabolism, immunity, brain function, and response to medical drugs. Our research objective is to mechanistically understand these host-microbiota interactions at the molecular level by addressing the fundamental question of how microbes recognise and transform their chemical environment.
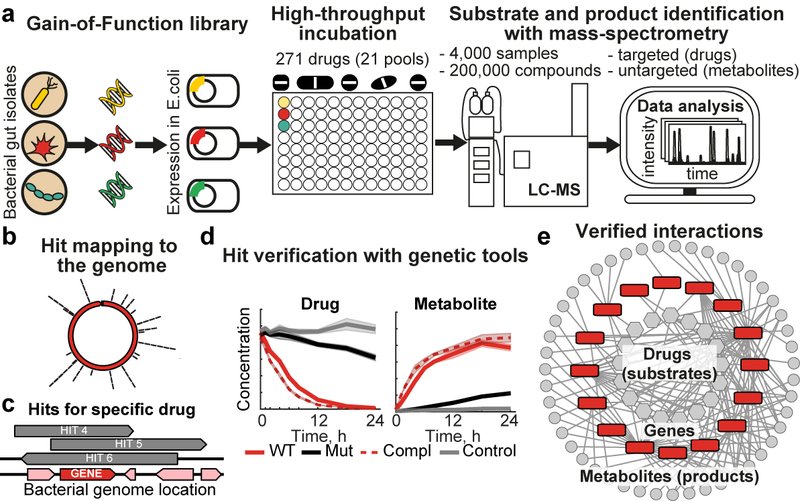
Bacterial enzymes determine the metabolic capacity of a microbial community, hence knowing their encoding genes is essential for a functional understanding of the microbiome. We have developed systematic approaches that combine high-throughput mass spectrometry, bacterial culturing and genetics to assay biotransformation of chemical compounds by gut commensals and to discover the responsible enzyme-encoding genes (Fig. 1). We are applying these experimental pipelines on medical drugs, nutrients, and environmental toxins, which improves the functional annotation of microbiome sequencing data and thus our understanding of the metabolic consequences of the variability in microbiome composition.
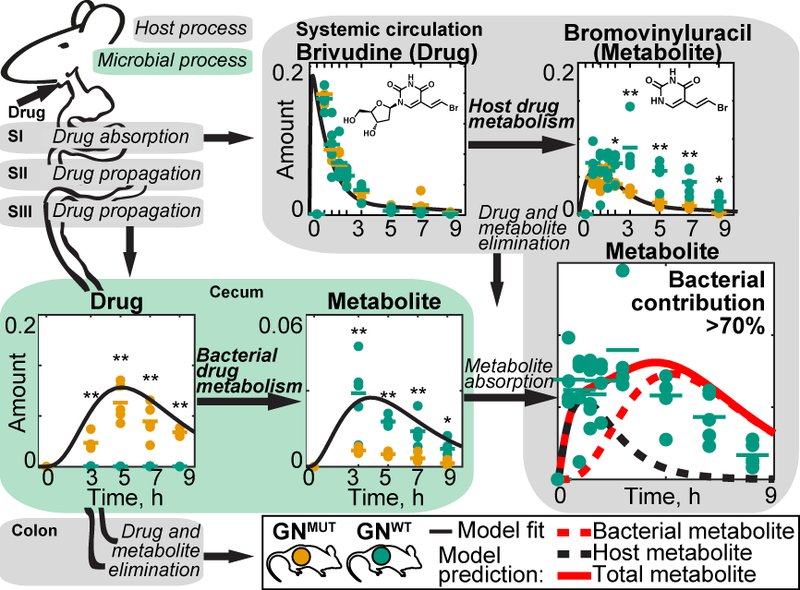
To understand the metabolic interplay between a host and its gut microbiota, it is necessary to quantify metabolite transfers between the two partners. Since drug molecules resemble endogenous metabolites, can be metabolised, but are not produced in vivo, they are ideal probes to investigate such metabolite transfers. We combine genetic manipulation of human commensals with gnotobiotic mouse models to quantify the impact of microbial biotransformation in the gut to drug and metabolite kinetics in circulation and host tissues. Informed by these measurements, we build mathematical models to quantify and predict compound exchange between microbiota and host. Such in silico models predict how physiological, microbial and chemical parameters determine metabolic host-microbiome interactions in the context of xenobiotic metabolism and more generally under physiological (non-drug) conditions (Fig. 2).
We aim at expanding our data on microbial biotransformation to: