Alejandro Torres-Sánchez
Group Leader
ORCID: 0000-0002-4275-173X
EditComputational modelling of multicellular systems

Group Leader
ORCID: 0000-0002-4275-173X
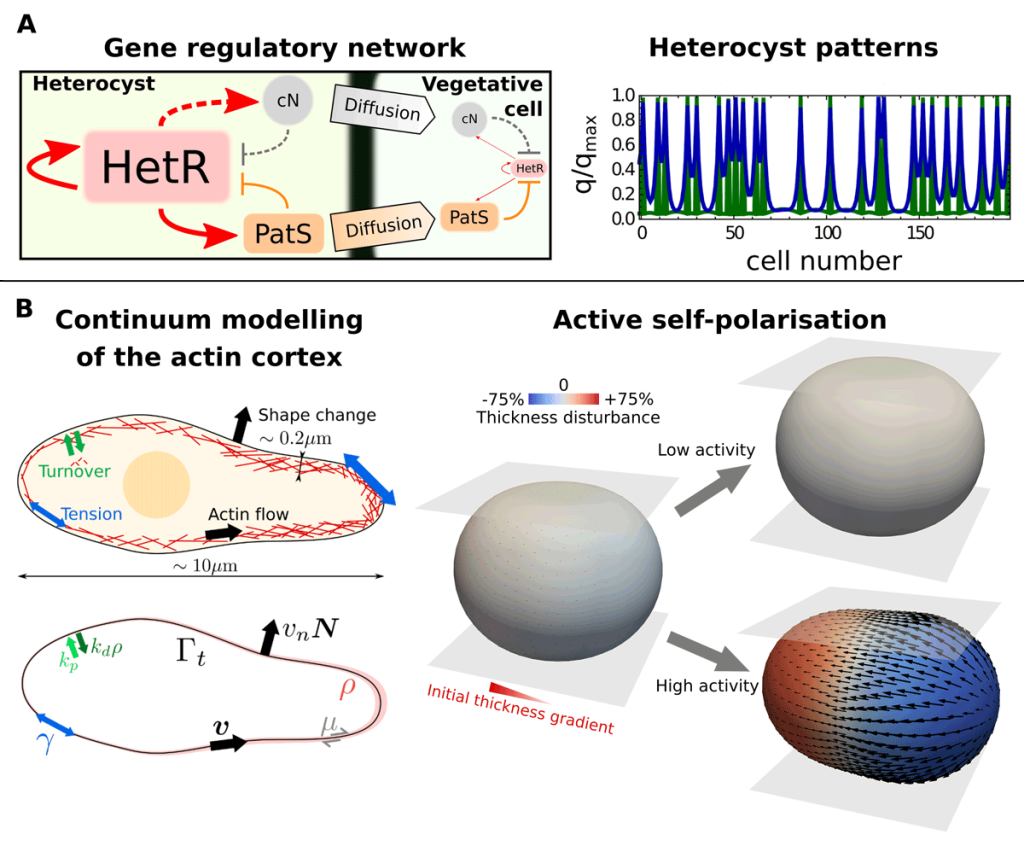
EditOur work lies at the intersection between theoretical physics, computational engineering, and quantitative biology. We develop mathematical models to understand the fundamental principles behind tissue self-organisation and shaping, both in embryos and in vitro tissue models like organoids. These include, for instance, dynamical-systems models to explain cell differentiation and tissue patterning and active-matter models to describe the mechanics of cells and tissues (see Figure 1 and Video 1). To understand these models beyond linearised solutions in simple geometries, we develop numerical methods and high-performance computer codes. Computational methods are a fundamental tool to solve mathematical models of biological processes as these typically involve nonlinear interactions of their constituents that are difficult to fully capture with linearised models. Numerical methods are also important for a faithful comparison of theoretical predictions with experiments, which often involve complex geometries and topologies that cannot be treated analytically.
During the breathtaking process of embryo development and organ formation, a single cell generates a complex organism through an interplay between cell proliferation, cell-fate decisions, mechanics, and biochemical patterning. How does this interplay lead to the complex and precise shapes and organisations of embryos? And how do failures in these interactions lead to disease? The ultimate goal of our research is to help answer these questions, together with experimental collaborators, using tools from theoretical physics and computer simulations.
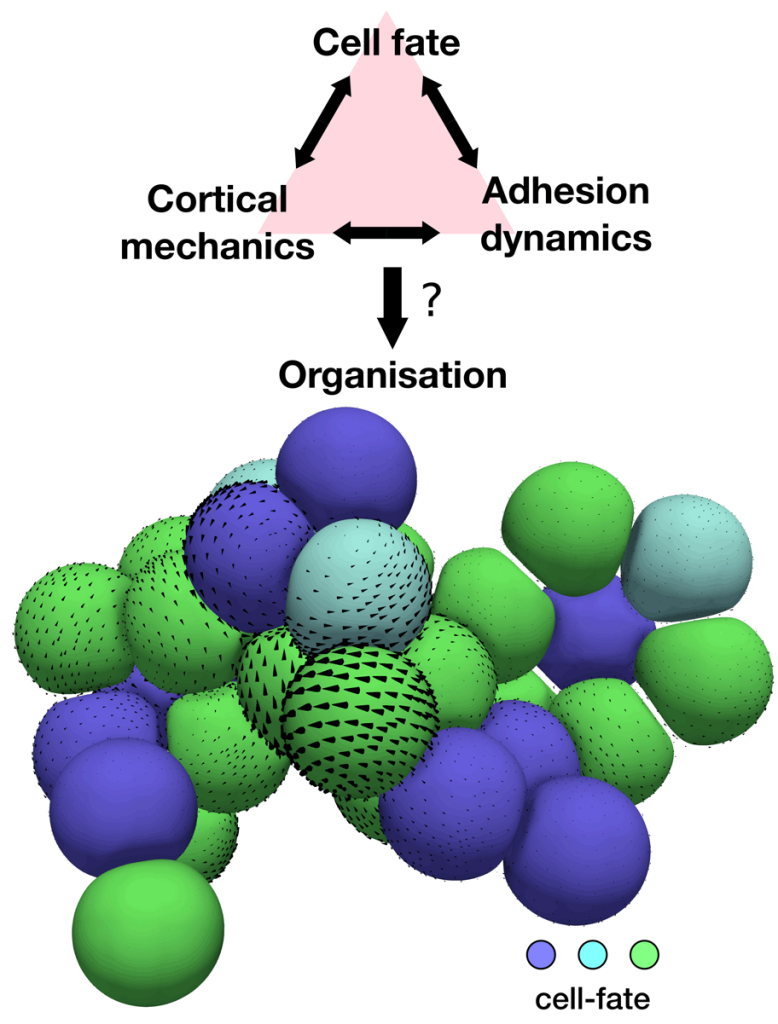
Different active tissue models, such as vertex, cell-based, or continuum models, have successfully been applied to understand morphogenetic processes. Despite this success, there are still gaps in our current understanding of morphogenesis that require new theoretical approaches. For instance, we would like to tackle questions such as, what interactions between mechanics and cell fate decisions are required to acquire a given tissue organisation and shape? Or, how do microscopic details, such as the distribution of the actin cortex or the dynamics of cell-cell adhesions, control the organisation of cell aggregates? (See Figure 2). Filling these gaps is also important for the rational design of novel self-organising materials based on promising new technologies in the field of synthetic biology, such as in vitro organoids.