Georgia Rapti
Group Leader (Outgoing)
ORCID: 0000-0003-4836-8640
EditAssembly mechanisms of nervous system architecture

Group Leader (Outgoing)
ORCID: 0000-0003-4836-8640
EditThroughout an embryos’ life, cells with diverse fates and morphologies interact to give rise to well-defined tissue architecture. The early assembly of nervous system circuits is fundamental for life and behavior; yet dissecting the events initiating this assembly is a tantalising challenge. How do nervous system cells interact spatio-temporally while concurrently diversifying their fates? Pioneer cells are thought to initiate circuit assembly, but their molecular identity, development, and interactions remain elusive. How do circuit components establish their specialised morphologies to coordinate assembly in vivo, towards functional connectivity? What interactions ensure the fidelity with which the mature nervous system structure is maintained? Which overarching principles pattern assembly across species?
To address these questions, we employ an interdisciplinary line of real-time in vivo imaging and refined genetics, genomics, and molecular approaches. We will combine cellular and molecular studies with high-resolution imaging and omics approaches, in collaboration with fellow research groups. The main system we study is the nematode C. elegans. This is a valuable system for our interdisciplinary approach, since C. elegans has transparent embryos, traceable lineages, morphogenesis that is tractable at single-cell resolution, mapped nervous system anatomy and connectivity, a sequenced genome, and sophisticated genetics (see figure and videos). Many C. elegans neural cell types and genes are shared across invertebrate and vertebrate species. Thus, discovery of gene–function relationships and the dissection of nervous system assembly in C. elegans is our route to addressing – in collaboration with other research groups – conserved principles in other organisms.
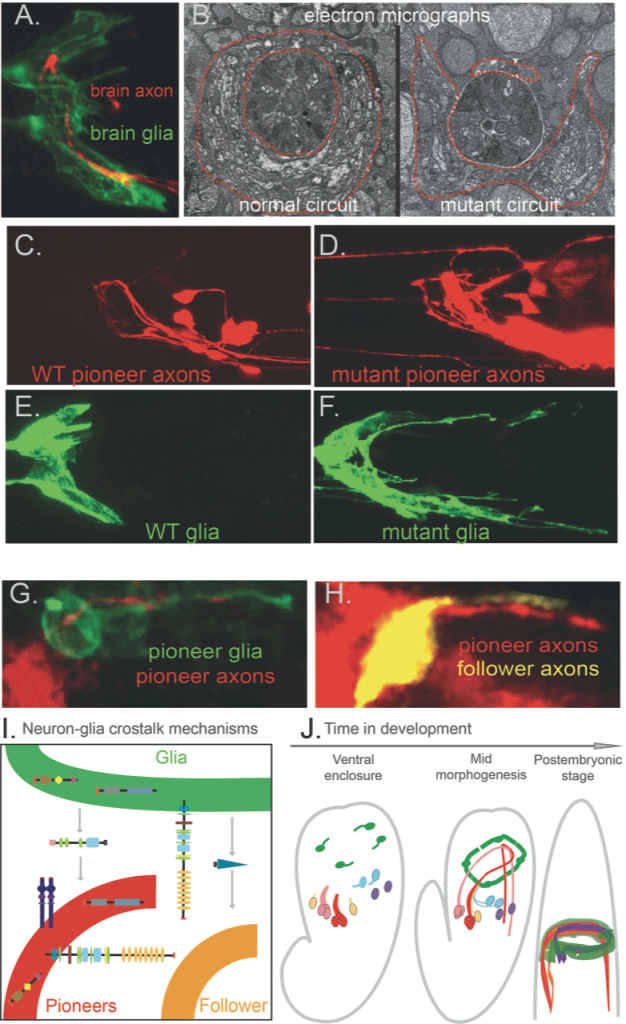
Centralised nervous systems consist mainly of neurons and glia – lineally related cell types in about equal numbers. Neurons transmit electrical currents, while glia were long thought to passively support neurons’ nutrition. However, glia were recently implicated in nervous system development and function, in physiology and pathology. We identified that C. elegans glia, similar to vertebrate glia, initiate hierarchical brain assembly: pioneer neurons and glia of specific molecular signature that cooperate functionally drive circuit assembly, they grow coalescing processes and molecularly guide circuit components (Figure 1). A set of diverse, conserved molecular cues drive circuit assembly synergistically through a glia–neuron crosstalk that we are only beginning to understand. To dissect dynamic circuit assembly at the cellular and molecular levels, we investigate normal and defective development of the nervous system from the level of single neurons or glial cells to the scale of multicellular circuits (Fig.1). We can then trace back the emergence of circuits in real time (Figure 1, movies 1-2).
We map molecular and cellular interactions driving circuit architecture in vivo by dissecting: