Christoph Müller
Group Leader and Senior Scientist
ORCID: 0000-0003-2176-8337
EditMolecular mechanisms of transcriptional regulation in eukaryotes

Group Leader and Senior Scientist
ORCID: 0000-0003-2176-8337
EditWe study how sequence-specific transcription factors assemble on DNA and how they interact with co-activators and general transcription factors to recruit RNA polymerases to their transcription start sites. We are also interested in the overall structure, architecture and the inner workings of large molecular machines involved in the transcription process, such as RNA polymerases I and III, their specific pre-initiation complexes and chromatin-modifying complexes. Finally, we would like to gain insight into how DNA sequence information and epigenetic modifications work together to regulate gene transcription. To achieve these goals, we use structural information obtained by cryo-electron microscopy and X-ray crystallography, combined with other biophysical and biochemical approaches.
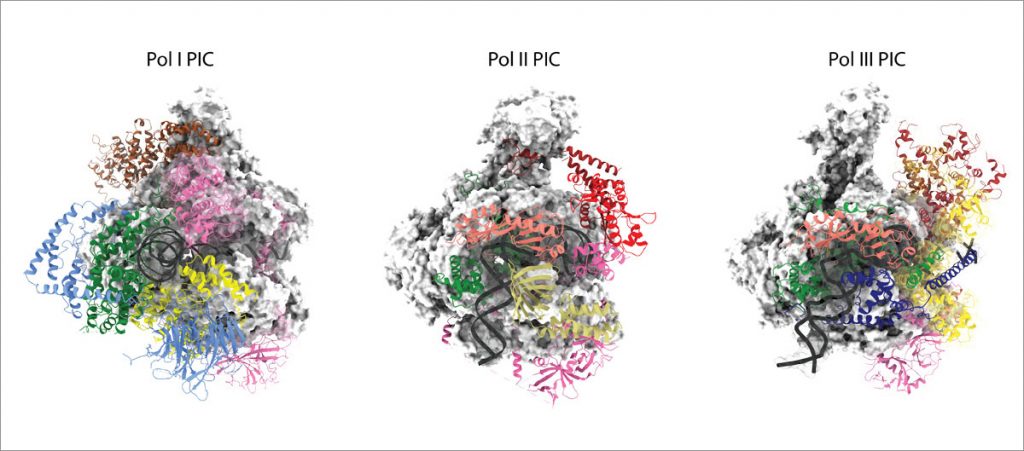
RNA polymerases I (Pol I) and III (Pol III) consist of 14 and 17 subunits, respectively. Whereas Pol I is responsible for the biosynthesis of ribosomal RNA, Pol III synthesises small RNAs like tRNA and 5S RNA. Misregulation of Pol I and Pol III has been associated with different types of cancer. We are studying the overall architecture of the Pol I and Pol III enzymes and of their pre-initiation machineries using a broad and interdisciplinary approach that combines integrated structural biology with in vitro and in vivo functional analysis. Analysing the Pol I and Pol III transcription process in detail will help us to understand what features make the Pol I and Pol III machineries particularly suitable to fulfil their respective tasks, how Pol I and Pol III transcription is regulated, and how Pol I and Pol III transcripts are further processed.
Pol I transcription takes place in the nucleolus, which is the site of ribosome biogenesis. The formation of the nucleolus is associated with Pol I transcription. The Pol III-specific general transcription factor TFIIIC can also act as an insulator in genome organisation. To locate and better understand the role of Pol I and Pol III transcription complexes in situ, our group uses cryo-electron tomography (cryo-ET) and correlative light and electron microscopy (CLEM).