Arnaud Krebs
Group Leader
ORCID: 0000-0001-7999-6127
EditDecoding gene regulation using single-molecule genomics

Group Leader
ORCID: 0000-0001-7999-6127
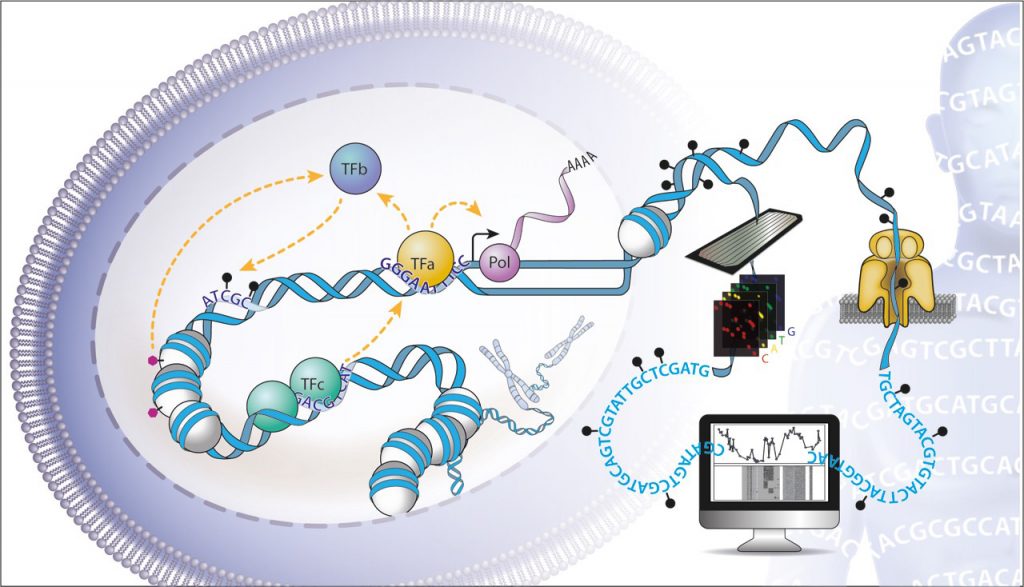
EditAbout 7% of the three billion bases contained in our genomes encode cis-regulatory elements, which control the activity of genes. Transcription factors interpret this genetic information to create the >250 different cellular identities required to form a multicellular organism. Expression of a gene is controlled by the collective action of multiple transcription factors, which recruit cofactors that alter chromatin, reshape genome organisation, and initiate transcription. The aim of our research is to understand how these multiple layers of regulatory information, and the factors that interpret them, integrate to precisely define gene expression levels (Fig. 1). How do Transcription factors cooperate? How is their collective function encoded in our genomes? What is the influence of epigenetics on their action? How do they communicate signals across large genomic distances? To address these questions, we develop high-throughput experimental and computational strategies to dissect the function of the regulatory genome.
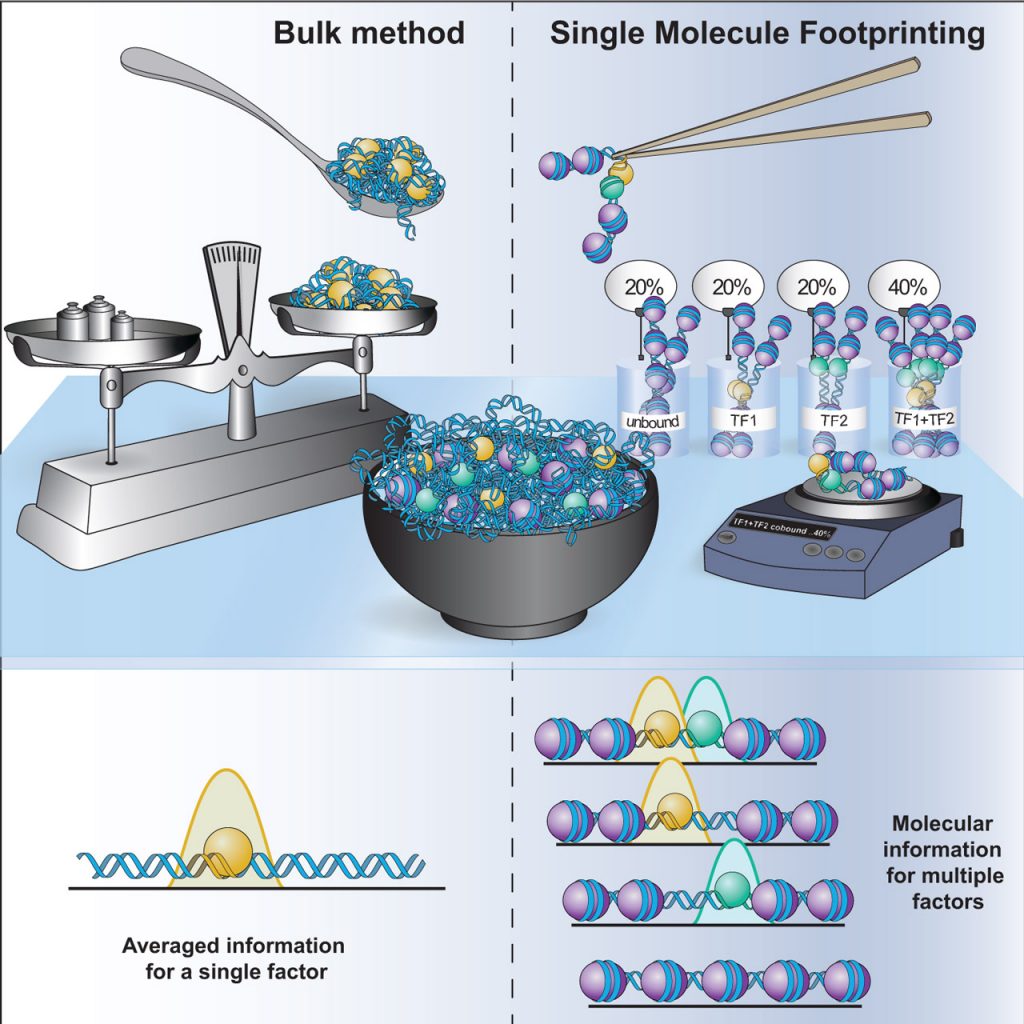
Single Molecule genomics: We developed protocols (Kleinendorst R, Barzaghi G, et al Nature Protocols, 2021) and computational tools for single-molecule footprinting (SMF), a methodology to quantify protein–DNA contacts at the level of single DNA molecules in vivo. SMF overcomes some of the limitations of bulk and single-cell genomics assays to quantify the patterns of occupancy of transcription factors at regulatory regions (Krebs A, Trends in Genetics, 2021).
Transcription factor cooperativity: Cooperativity between transcription factors is widespread at regulatory elements, but the underlying mechanisms are mostly unknown. We have analysed the molecular co-occupancy of transcription factors genome-wide and revealed principles ruling their combinatorial action at regulatory regions (Sönmezer C et al, Mol Cell, 2021) (Fig. 2).
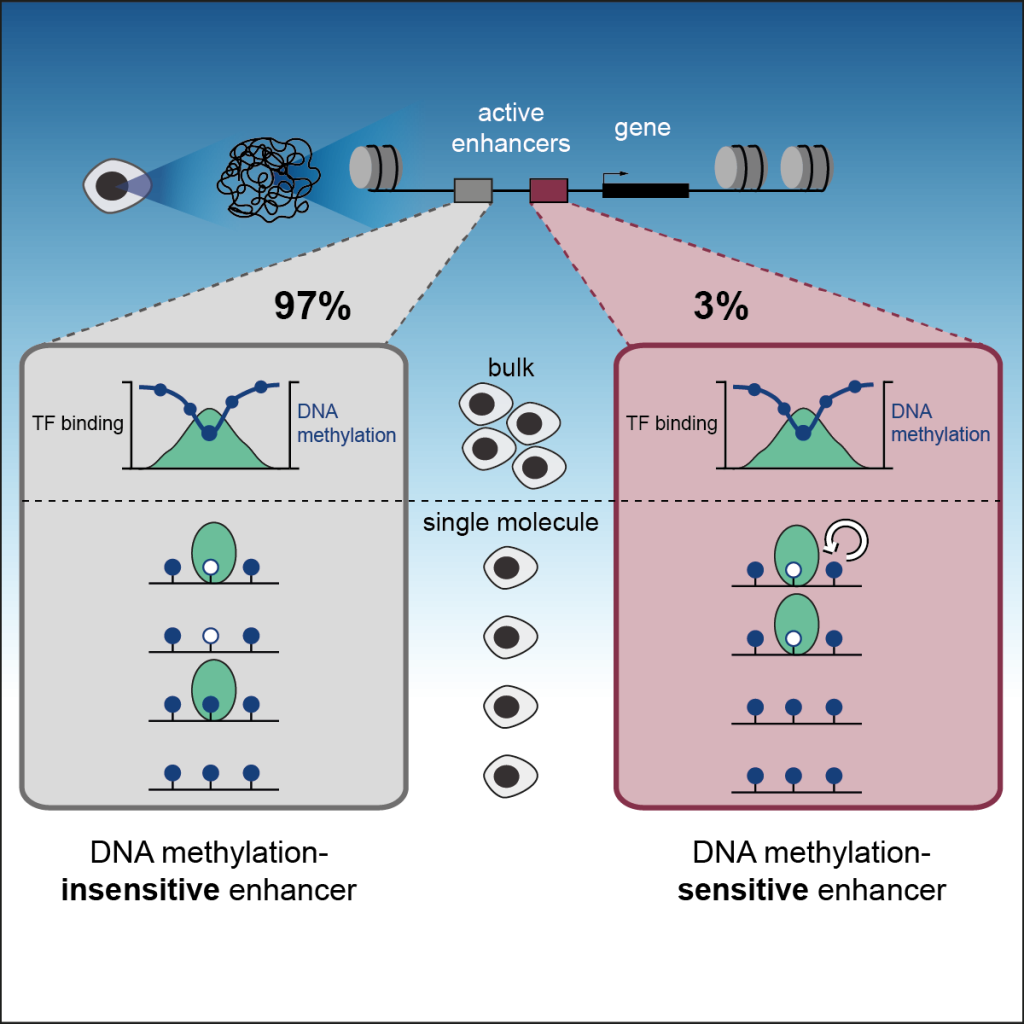
Epigenetic control of transcription: Epigenetic modifications and transcription factors mutually influence each other to control genes during development. We developed a strategy to measure how the presence of DNA methylation inhibits the binding of TFs on individual DNA molecules in vivo. This revealed the contribution of DNA methylation to the regulation of enhancer activity, the regulatory elements that control cell identities (Kreibich E et al, Mol Cell, 2023) (Fig. 3).
In the upcoming years, we will develop experimental strategies to perturb the function of transcription factors and epigenetic regulators at a scale and a resolution compatible with the understanding of the general principles of their function in the transcription process. We will further extend the single molecule genomics toolbox to monitor the effects of these perturbations on multiple regulatory layers across entire cis-regulatory units. We will apply this paradigm to understand how transcription factors interact with various chromatin contexts, and the genetic encoding of their function. We will use chemical and genetic perturbations to establish the causality and the directionality of the interactions between regulatory factors. We will undertake a biophysics approach to derive quantitative models able to predict the collective function of transcription factors in the context of chromatin. Understanding these general principles has potential applications in various fields such as the prediction of the functional impact of genetic variation across individuals or the rational design of cell-type specific cis-regulatory units for gene targeting.