
Jan Kosinski
Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-3641-0322
EditIn-cell structural systems biology of infection

Group Leader and Co-chair of Infection Biology Transversal Theme
ORCID: 0000-0002-3641-0322
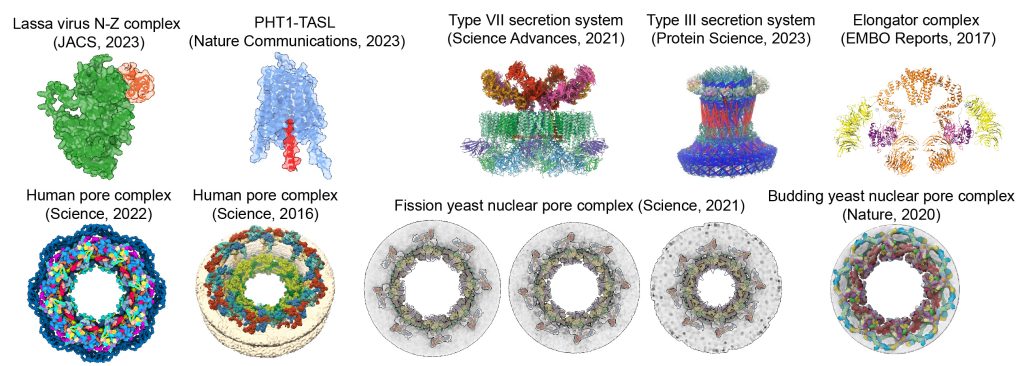
EditWe develop and apply computational methods for modeling macromolecular complexes by integrating AI-based structure prediction, electron microscopy, crosslinking, and other data. We have built models of one of the largest complexes in the cell – the nuclear pore complex from humans (Nature, 2015; Science, 2016, Science, 2022) and other species (Nature Communications, 2018; Nature, 2020; Science, 2021). We also collaborate on other complexes (e.g., EMBO Reports, 2017, Science Advances, 2019, Protein Science, 2023; Nature Communications, 2023; JACS, 2023) and membrane complexes involved in malaria (Plasmofraction project). Our modeling and analysis methods are available in various software packages, including Assembline for integrative modeling, AlphaPulldown for AI-based high-throughput structure prediction and interaction mapping, and AF3x for AlphaFold 3-based crosslink-driven modeling.
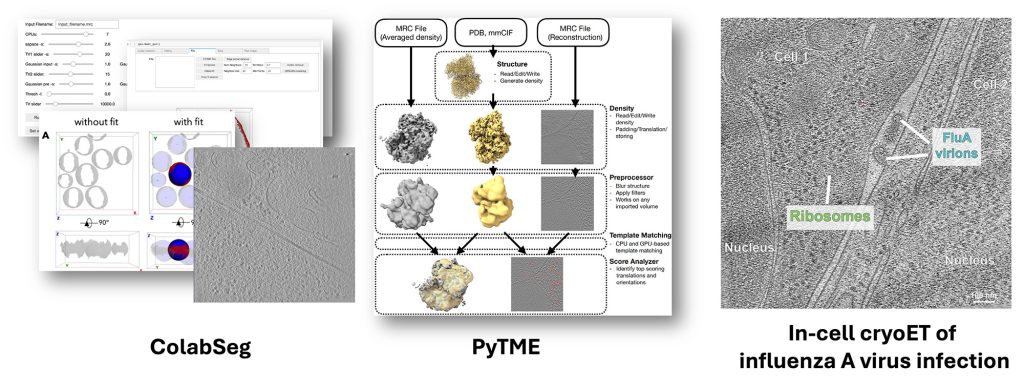
We develop and apply methods for structural biology in cells, primarily based on cryogenic electron tomography (cryo-ET) data. Our methods include ColabSeg software for membrane segmentation (now superseded by Mosaic), PyTME, an ultrafast and flexible template-matching engine, and HMFF and Mosaic for membrane segmentation, membrane modeling, simulation, and morphometrics. We also apply in-cell cryo-ET to study influenza A virus infection and the cell biology of the Giardia lamblia parasite. Within our recent ERC Synergy Grant, we will push the development of modeling and cryo-ET analysis methods to fundamentally redefine in-cell cryo-ET data mining as an integrative structural modeling problem.
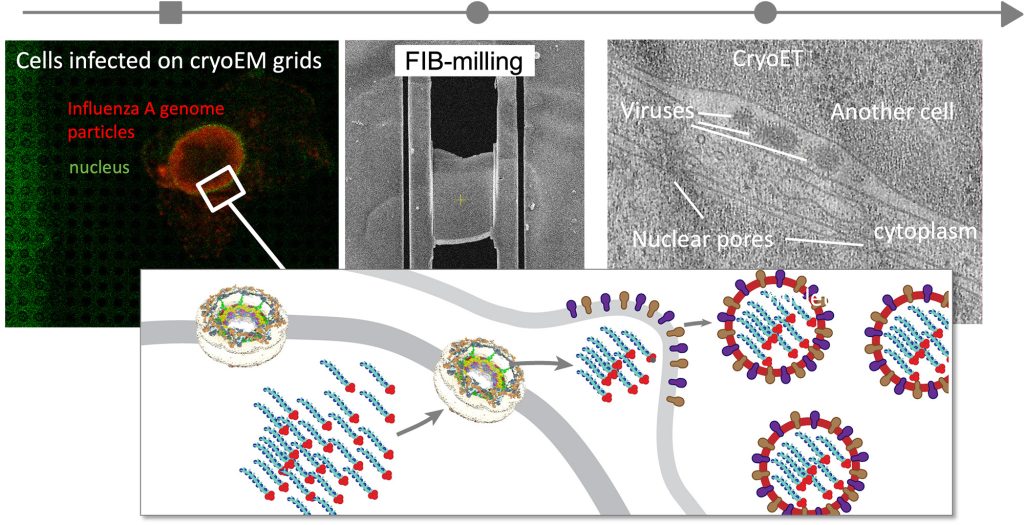
During infection, viruses undergo complex life cycles, interact with the molecular systems of their hosts, and disturb and hijack host molecular machines for their own purposes. We aim to create comprehensive multiscale models of entire infection cycles to discover host-pathogen interactions and identify which of them are the most crucial for infection. To this end, we integrate proteomics, advanced fluorescence microscopy, and cryo-electron tomography. Currently, we focus on the influenza A virus in our own lab, recently obtaining the first in-cell structural snapshot of influenza virus host-pathogen interactions.
Our lab is located in the Centre for Structural Systems Biology (CSSB), right next to EMBL Hamburg on the DESY campus. We are an interdisciplinary team of scientists who combine computational biology (structural modeling, systems biology) with wet lab experimentation (cell biology, proteomics, advanced fluorescence microscopy, electron tomography). Our research is highly collaborative and involves joint projects with other EMBL and CSSB groups, CSSB partners, and external groups.
