
Elizabeth Duke
Team Leader
ORCID: 0000-0003-2048-9598
EditBiological X-ray imaging

Team Leader
ORCID: 0000-0003-2048-9598
EditThe Duke team is developing high throughput tomography (HiTT) with the goal of offering this as a user service on EMBL beamline
P14 at the PETRA III synchrotron in Hamburg. Here the well-established infrastructure developed over the past decades for Macromolecular Crystallography is efficiently utilised for x-ray tomography.
We successfully performed x-ray tomography acquisitions of various biological sample types using free propagation phase contrast x-ray imaging. In addition, we have embedded the x-ray imaging data collection and data reconstruction into the existing EMBL Hamburg computation environment bringing down the acquisition and processing times to a few minutes. The main goal of our work is to provide a very flexible but streamlined x-ray imaging setup capable of analysing vastly different sample types from small organisms to whole tissue samples in a standardised and streamlined fashion.
One of our particular interests is in imaging tissue samples from various origins at multiple length scales. While in preclinical research and histopathological clinical routine, tissue samples are commonly analysed by traditional sectioning based histology, we are expanding tissue analysis into the third dimension by performing so-called 3D virtual histology. We can image both tissue in liquid (hydrated and dehydrated) and also tissue that has been paraffin or resin embedded.
Currently each individual data set has a field of view of 660 um and a resolution of ~700 nm which means that we can see individual cells. As most tissues don’t come in convenient 660 um cubes we have developed the option to tile together adjacent data sets from a larger sample enabling us to build up composites of, for example 1 mm punch biopsy tissue cores.
We can also image samples that have been prepared for volume electron microscopy – i.e. heavy metal stained, resin embedded.
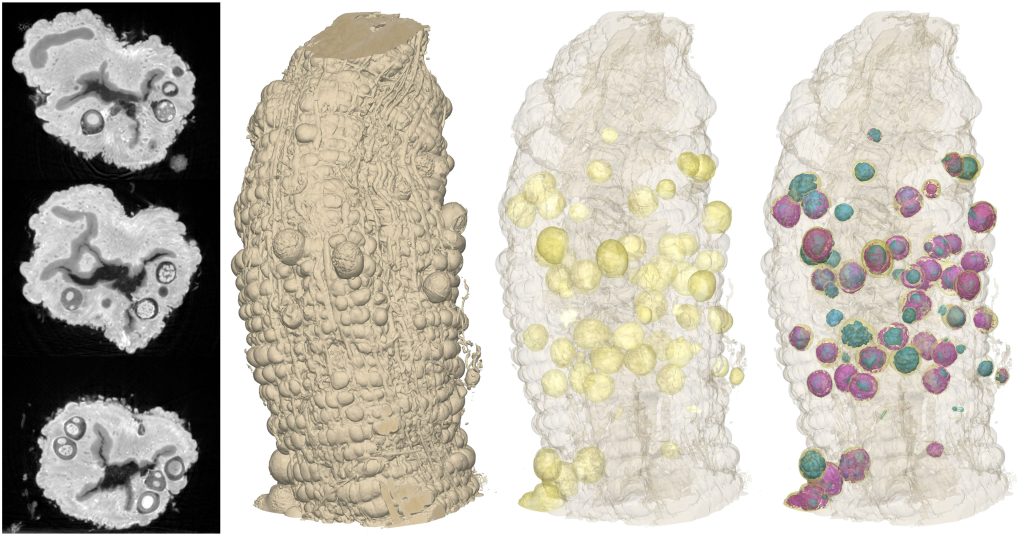
Because of the high throughput nature of our imaging technique we are able to image many samples in a short period of time. This opens the door to carrying out 3D population studies – in effect 4D X-ray imaging. One could use the technique to examine areas of developmental biology, looking at n-day old model systems.
Alternatively one could also use HiTT to image vast quantities of field collected samples in order to screen them for further more extensive downstream analysis.
X-ray imaging in general is incredibly useful as part of a correlative imaging pipeline. By that we mean imaging one sample with different modalities and then overlaying all the data – the final result is greater than the sum of the individual parts. This is particularly the case when used as part of the established correlative work flow linking light microscopy with volume electron microscopy. X-ray imaging allows us to, for example, zone in on a region of interest to enable much more efficient high resolution data collection.
The aspiration of the team is to enable hard X-ray imaging of soft biological tissue to be offered to users with minimal expertise in the details of the imaging itself, providing them with support and guidance in every part of the process. Underpinning the development of all aspects of the X-ray imaging user facility is the desire to develop a 3D histology platform for biological tissue complementing and extending what is routinely available in 2D histology in the clinic.
