Read the latest Issue
It’s boring, but we all have to do it: cleaning our homes, and throwing out the rubbish we produce every day. If we didn’t, our homes would become dirty, smelly, and a threat to our health.
Cells have to perform the same kinds of housecleaning tasks, as proteins and organelles such as mitochondria grow old, stop functioning properly and become toxic. If these components are not processed and removed, they can be fatal risk for the cell, as occurs in some neurodegenerative diseases. Now new research led by EMBL scientists, and published in Cell Reports, has shed new light on how cells go about taking out their rubbish.
Cells clear out their trash through a two-step process called autophagy. First, cellular debris is collected within bubble-like vesicles called autophagosomes. Next, these autophagosomes are delivered to lysosomes, membrane-bound sacs where the cell’s rubbish is broken down by specialised enzymes.
While the broad outlines of this process are well known, the precise details of how it happens have remained unclear. The new research led by Carsten Sachse, Group Leader in the Computational and Structural Unit at EMBL Heidelberg, focused on the role of a protein called p62, which plays an important role in autophagy by delivering cellular trash to autophagosomes.
Previous studies have revealed that p62 is made up of three domains, each with a different role. The PB1 domain enables single p62 proteins to join together to form long chains. Another, the UBA domain, recognises special protein tags called polyubiquitin that are attached to proteins and organelles that need removing. The third domain, LIR, enables p62 to bind to autophagosomes, which ensures that any cargo attached to UBA ends up inside the autophagosome.
Autophagosomes have to engulf big structures like mitochondria, so they need a scaffold like p62 filaments to grow sufficiently large
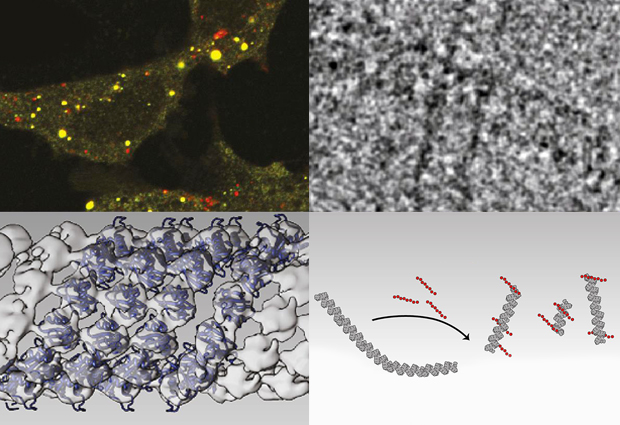
These earlier studies, however, looked at portions of p62 in the test tube, and not the entire native protein. To do so, Sachse and colleagues, including a team of cell biologists from the University of Tromsø, Norway, used cryo-electron microscopy to look at long chains of p62 as they are likely to exist in the cell. “CryoEM is well suited to looking at large structures like p62 polymers”, explains Sachse.
The images obtained with cryoEM were then combined with structural data from earlier work to create, for the first time, detailed 3D models of p62 polymers. The team found that p62 joins up to create long, organised filaments in which the p62 subunits are wound around a helical axis to from a tube with a hollow core. Within this helical filament, the p62 subunits are arranged so that the UBA and LIR domains face outward, and biochemical studies confirmed that these domains can interact with polyubiquitin and autophagsomes, respectively.
Sachse and colleagues suggest that these flexible but strong p62 filaments serve as a scaffold which the membranes of the growing autophagosome are built on. “Autophagosomes have to engulf big structures like mitochondria, so they need a scaffold like p62 filaments to grow sufficiently large,” says Sachse. Although the idea of p62 acting as a scaffold had been proposed before, no one knew what it looked like before. “Now we do,” smiles Sachse.
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive