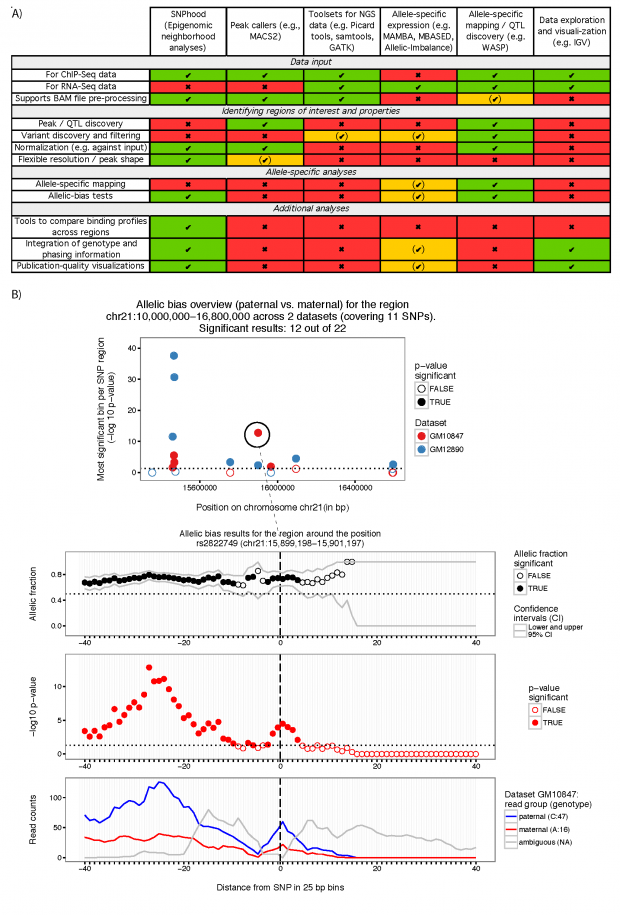
Motivation: The vast majority of the many thousands of disease-associated single nucleotide polymorphisms (SNPs) lie in the non-coding part of the genome. They are likely to affect regulatory elements, such as enhancers and promoters, rather than the function of a protein. To understand the molecular mechanisms underlying genetic diseases, it is therefore increasingly important to study the effect of a SNP on nearby molecular traits such as chromatin or transcription factor binding.
Results: We developed SNPhood, a user-friendly Bioconductor R package to investigate, quantify and visualize the local epigenetic neighbourhood of a set of SNPs in terms of chromatin marks or TF binding sites using data from NGS experiments.
Availability: SNPhood is publicly available and maintained as an R Bioconductor package at http://bioconductor.org/packages/SNPhood/.