Judith Zaugg
Group Leader
judith.zaugg [at] embl.de
ORCID: 0000-0001-8324-4040
EditSystems (epi)genetics to study the basis of complex traits and diseases

Group Leader
judith.zaugg [at] embl.de
ORCID: 0000-0001-8324-4040
EditThe Zaugg Group combines genetic and epigenetic variation across individuals to study molecular mechanisms underlying complex traits and diseases. They develop and apply single cell technologies and computational tools to address basic and translational questions focused on gene regulatory mechanisms.
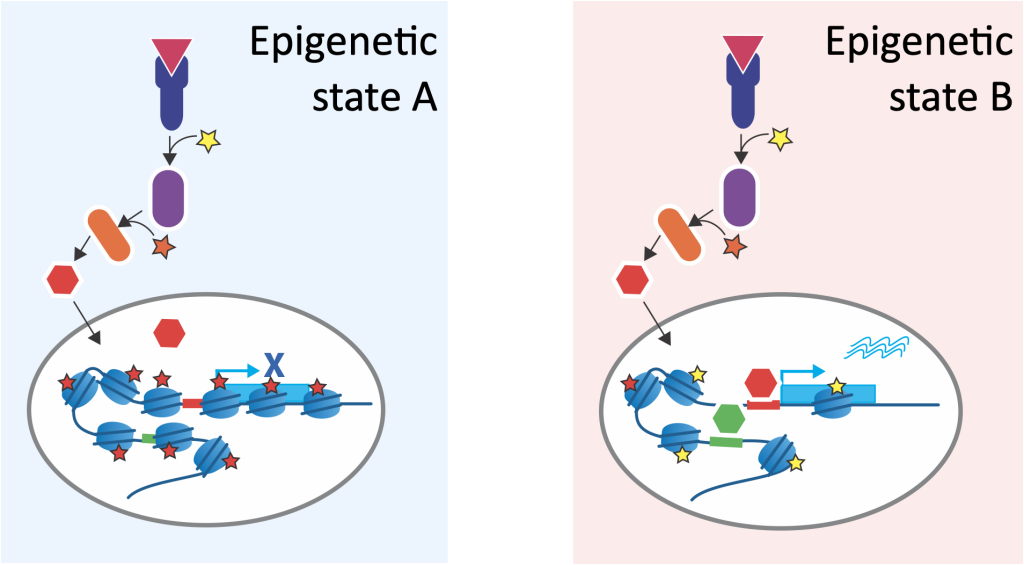
A crucial part of precision medicine is to quantitatively understand the interplay of genetics, epigenetics, and environmental factors, including the cellular microenvironment. Given that the majority of disease-associated genetic variants lay in non-coding regions, and environmental factors often leave an epigenetic signature, we believe that gene regulatory processes are key to understand disease mechanisms. Our research highlights the emerging role of gene regulation (including enhancers and transcription factors) in disease mechanisms.
Our research vision is to understand how cells integrate genetic information, epigenetic state, and extrinsic signals from their microenvironment, and how these layers give rise to cellular states that ultimately define complex (disease) phenotypes and response to perturbations.
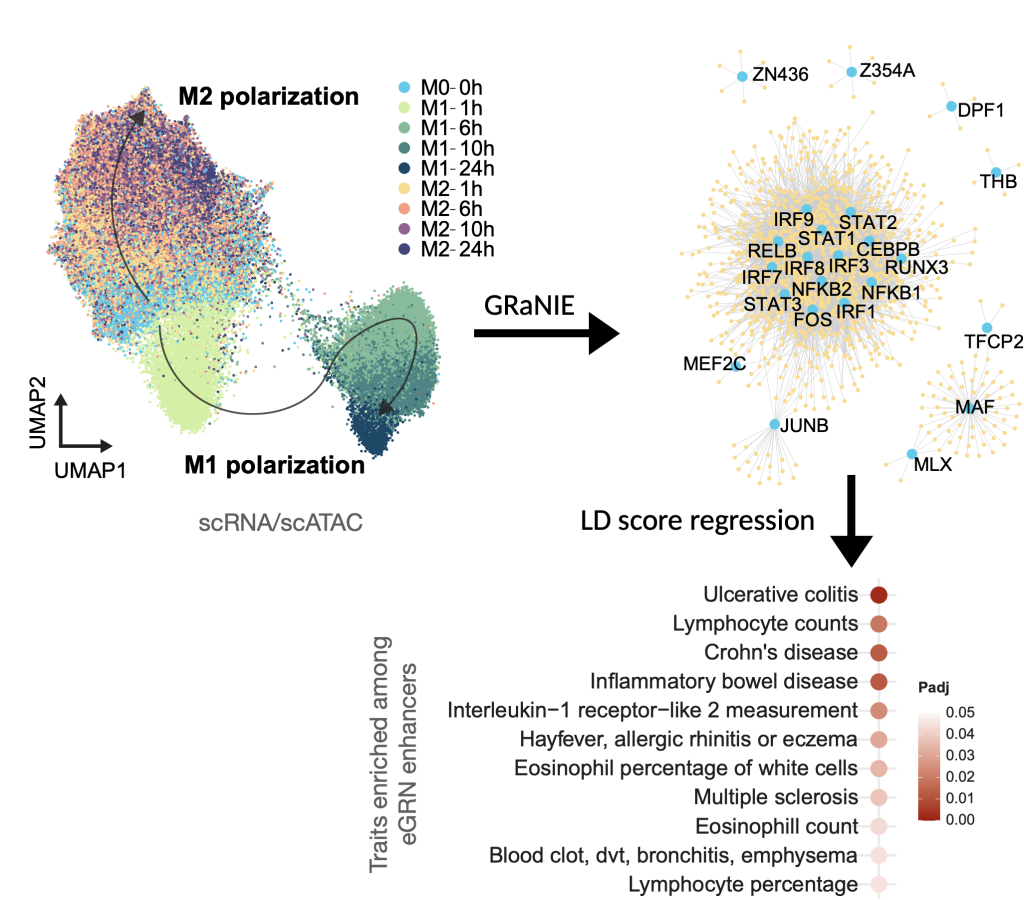
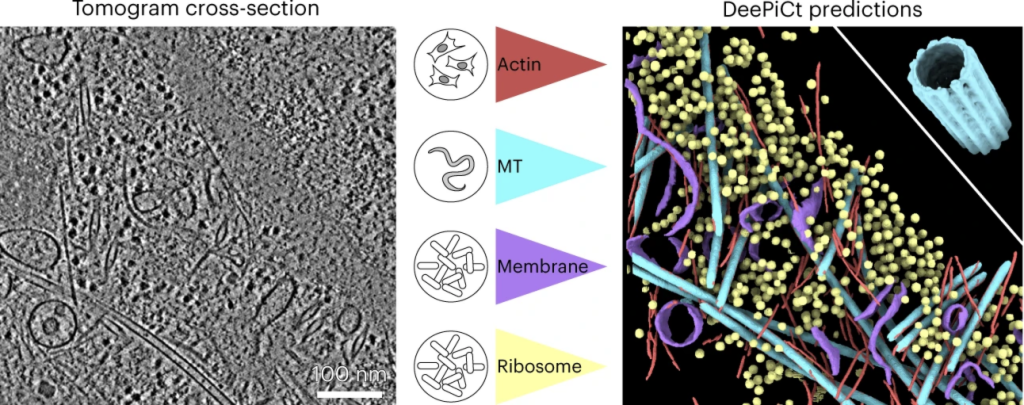
Our approach is to build systems biology frameworks for studying how cells respond to defined perturbations given their molecular and epigenetic profiles. For this we develop genomics technologies and computational tools to integrate data across scales and types, including single cell multiomics (RNA/ATAC), spatially resolved data and cryo-electron tomography.
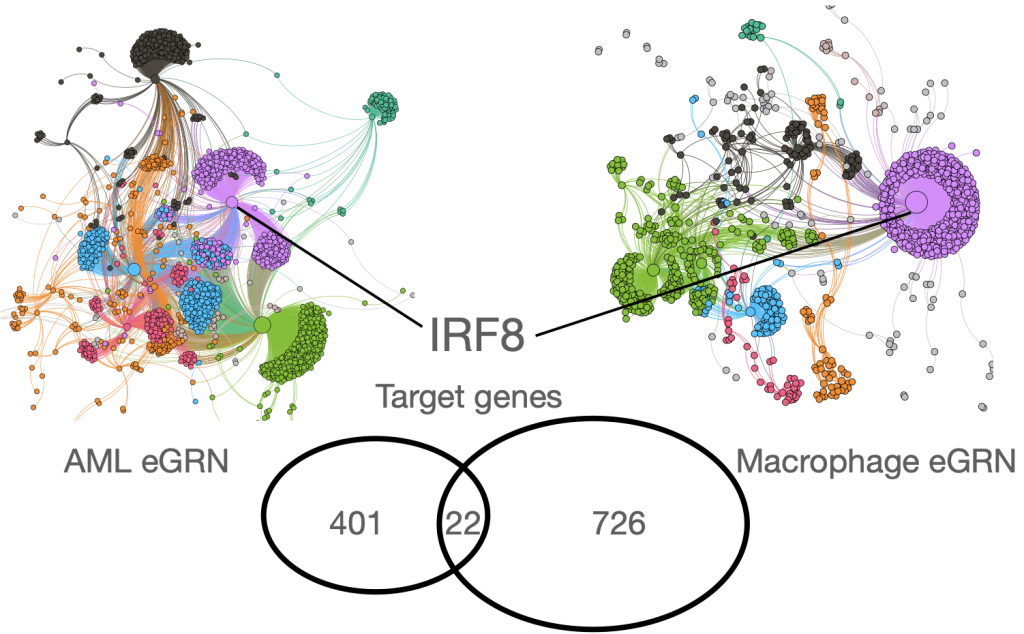
Our systems of interest are: (i) the bone marrow niche, where we aim to understand how stem and progenitor cells interact with their immune and stromal microenvironment, and how these interactions contribute to hematological malignancies and therapy response. (ii) immune cells and their interaction with tissue in immune system dysfunction, inflammation, infection, and cancer. Here we are interested in understanding the cell-type specific regulatory networks that contribute to disease mechanisms.
We are expanding our work on disease mechanisms, gene regulatory networks, and cell-cell interactions along three lines of research:
Examples of our methods and technologies and further reading:
Recent research highlights