Fabian Grubert*, Judith B Zaugg*, Maya Kasowski*, Oana Ursu*, Damek Spacek, Alicia R. Martin, Peyton Greenside, Rohith Srivas, Doug Phanstiel, Aleksandra Pekowska, Wolfgang Huber, Jonathan Pritchard, Carlos D. Bustamante, Lars M. Steinmetz, Anshul Kundaje, and Michael Snyder
* These authors contributed equally to this study.
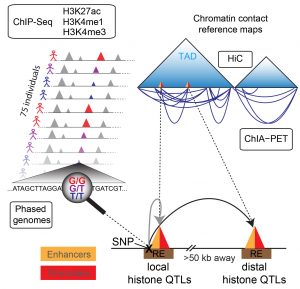
Deciphering the impact of genetic variants on gene regulation is fundamental to understanding the genetic and molecular basis of human disease. Although gene regulation often involves long-range regulatory interactions, it is unknown to what extent non-coding genetic variants influence distal molecular phenotypes. Here, we integrate chromatin profiling experiments for three promoter/enhancer associated histone marks in lymphoblastoid cell lines (LCLs) from 75 sequenced individuals with LCL-specific Hi-C and ChIA-PET-based reference chromatin contact maps to uncover one of the largest collections of local and distal histone quantitative trait loci (hQTLs) in a single cell type. hQTLs are mediated by extensive networks of chromatin contacts involving not only canonical enhancer-promoter interactions but also enhancer-enhancer and promoter-promoter associations. The associated elements predominantly lie within topologically associated domains and exhibit largely concordant variation of chromatin state coordinated by proximal and distal non-coding genetic variants that often disrupt binding sites of key lymphoid transcription factors. The hQTLs are enriched for common variants associated with autoimmune diseases and enable identification of putative target genes of disease-associated variants from genome-wide association studies. Our study thus provides novel insights into how genetic variation can affect human disease phenotypes by disrupting regulatory networks mediated by three-dimensional long-range interactions.
Data files:
Local histone QTLs, Distal histone QTLs, Additional files [For the code used in the analysis please contact Judith]