Ana Boskovic
Group Leader
ORCID: 0000-0003-1258-4376
EditMechanisms of embryonic gene regulation

Group Leader
ORCID: 0000-0003-1258-4376
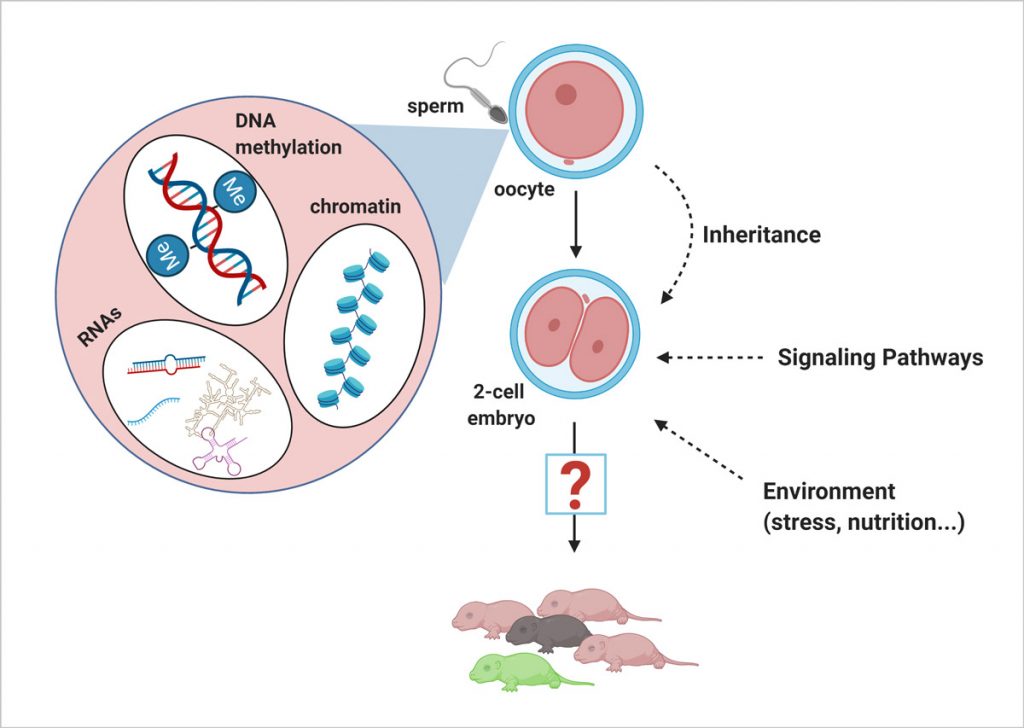
EditAt fertilisation, the oocyte and the sperm fuse to form the totipotent zygote, which will give rise to all other cell types in the developing organism. After gamete fusion, genome-wide epigenetic remodelling takes place on both parental chromosomes, ensuring the erasure of distinctive gamete-specific genome organisation. Newly formed embryonic chromatin is then subject to regulation by multiple waves of signalling pathways, metabolites, and epigenetic factors. These events, which together shape the transcriptome during embryogenesis, have the potential to affect all cell types in an organism.
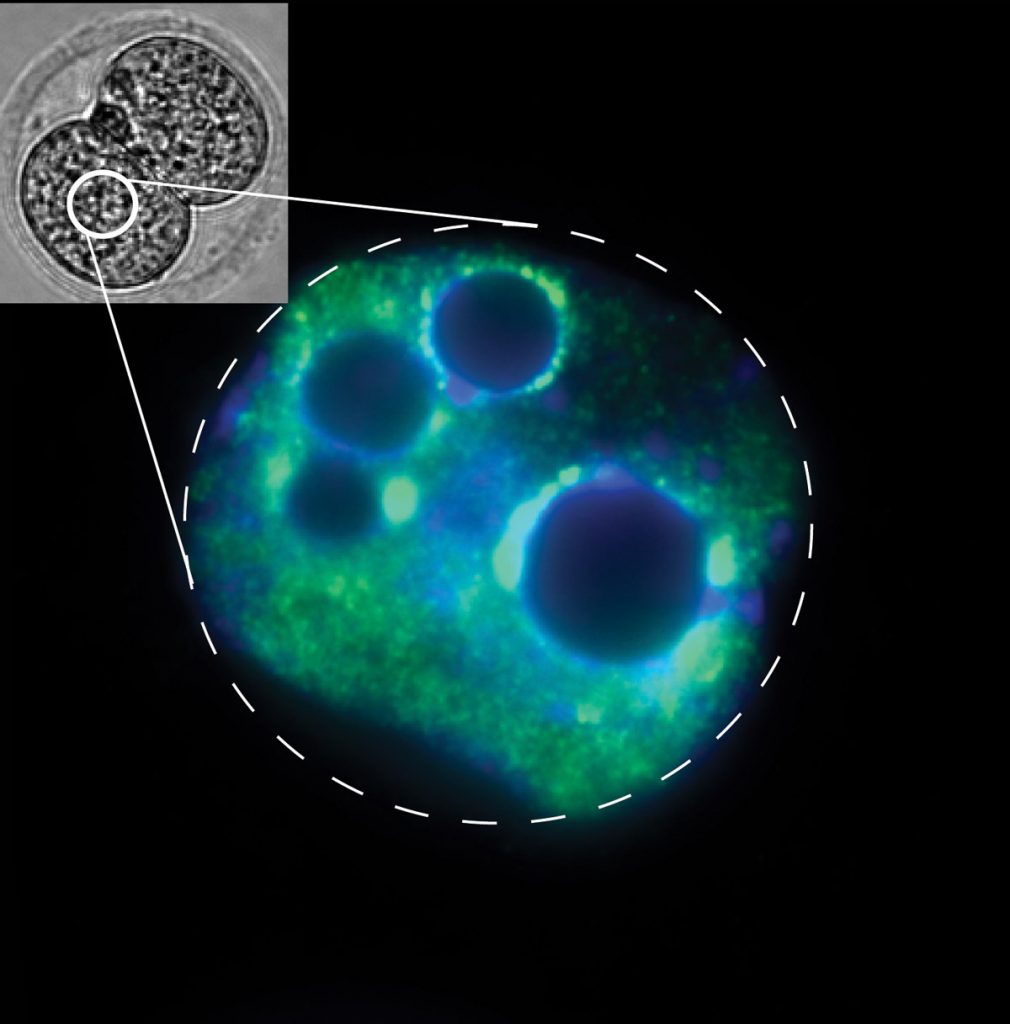
In the past decade, it has become increasingly well accepted that ancestral environments can influence epigenetic information that is heritable across generations. At present, however, there are only sparse molecular details about how phenotypic changes in the developing animal result from epigenetic factors delivered in the gametes, or from other environmental factors that the embryo experiences during a discrete developmental window. We have previously shown that, in mice, sperm-borne tRNA fragments responsive to paternal diet can influence the transcription of totipotency-associated genes through regulation of histone levels and chromatin organisation. Our lab is thus focused on gaining in-depth insight into mechanistic transactions regulating mammalian embryonic chromatin dynamics and gene expression, which is crucial for understanding how cellular identity is acquired, maintained, and regulated as cells differentiate during normal development.
The goal of our lab is to understand the mechanisms governing the establishment of gene expression patterns during mouse development, and how their modulation through epigenetic pathways influences early cell fate decisions and downstream developmental trajectories. Through a combination of cutting-edge genomics, molecular embryology, mouse ES cell manipulations, and traditional gain- and loss-of-function approaches, we aim to: