Janosch Hennig
Visiting Group Leader (Outgoing)
ORCID: 0000-0001-5214-7002
EditIntegrated structural biology of translation regulation mechanisms

Visiting Group Leader (Outgoing)
ORCID: 0000-0001-5214-7002
EditDosage compensation is an essential molecular process in sexually reproducing organisms, which compensates the imbalance of number of sex chromosomes between the sexes. Although these processes can be quite different between species, recent research shows that all have common mechanisms. One of which is the involvement of complexes between proteins and different RNA molecules, mRNA and long non-coding RNAs, often involved in phase separation.
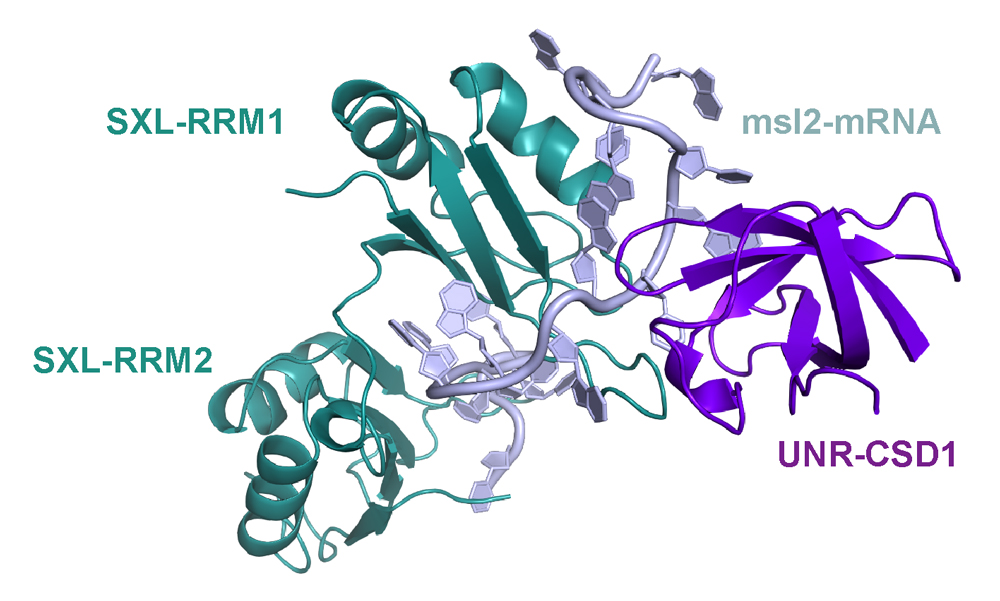
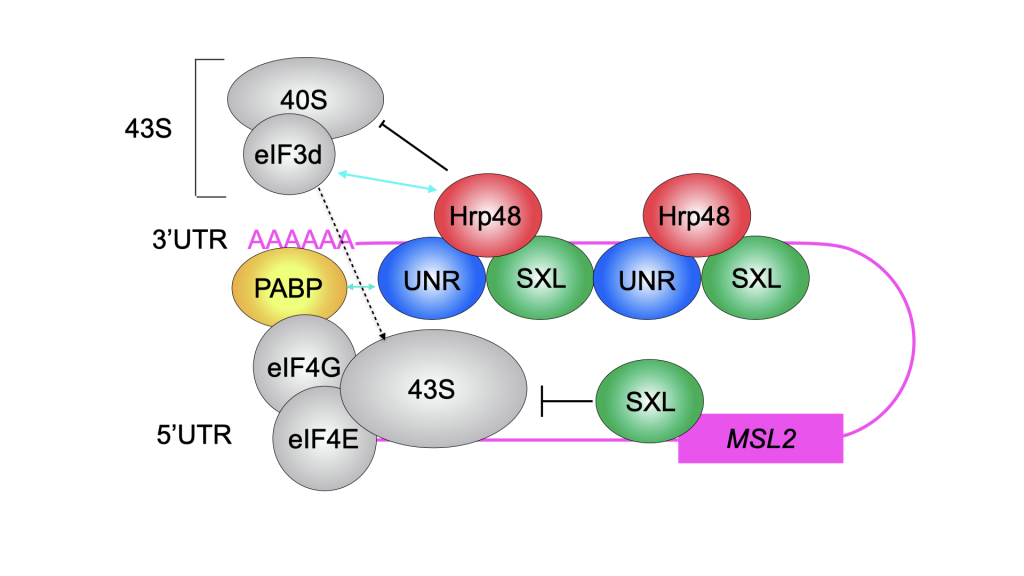
In Drosophila, we study both, the female and male side. In males, we want to understand how exactly the long non-coding RNAs RoX1 and RoX2 are remodelled to allow assembly of the dosage compensation complex (or MSL complex) on the single male X chromosome to achieve 2-fold hypertranscription. This hypertranscription would be lethal in females. Instead, the female-specific protein sex-lethal (Sxl) binds to the mRNA of the MSL complex’ rate-limiting component MSL2, to prevent its translation. The highly conserved protein Upstream-of-N-Ras (Unr) is recruited by Sxl to the same site on the mRNA (Figure1) and, together with Hrp48, essential for translation repression of msl2 mRNA (Figure 2). The male MSL complex is also conserved in humans, where it is regulating autosomal compensation. All RNA binding proteins regulating translation in female flies are conserved in humans, and Unr for example is highly expressed in certain cancer cell lines.
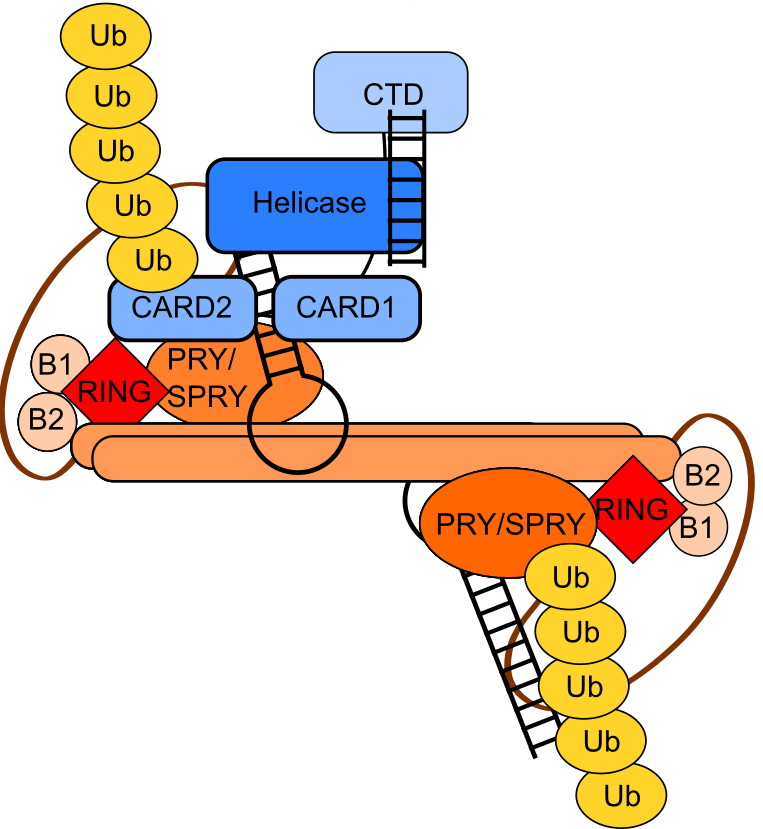
Another main project of the lab revolves around novel RNA binding proteins, meaning proteins, which do not feature a classical RNA binding domain (like RRM, CSD, KH or dsRBD domains), but have been shown to bind single-stranded RNA in mRNA interactome capture. Of special interest to us are RNA binding E3 ligases of the tripartite motif (TRIM) protein family. Here we want to understand how RNA binding is connected to these proteins’ main biochemical function: ubiquitination. Based on our data, we hypothesize that some TRIMs bind to mRNA and regulate translation by ubiquitinating components of translation complexes. Other TRIM proteins and their ubiquitination function seems to be actually regulated by RNA, which we could recently show for TRIM25 (Haubrich et al., BioRxiv, 2020, Figure 3).
Our ultimate goals are to obtain high resolution structures of these large protein-RNA complexes validated by biochemical and cell biological experiments to get a detailed molecular understanding of these essential mechanism. To this end, we employ all available structural biology methods (NMR, X-ray crystallography, cryo-EM and small-angle scattering) and more.
We also collaborate on many exciting projects within and outside of the EMBL, where we help out with our NMR and integrative structural biology expertise.