Justin Michael Crocker
Group Leader
ORCID: 0000-0002-5113-0476
EditGene regulation during evolution and development

Group Leader
ORCID: 0000-0002-5113-0476
EditThe goal of my research group is to reach a level of understanding that will allow the programming and control of developmental fates, extending existing single-cell synthetic biology techniques to whole organism developmental systems. Development is controlled, to a large degree, by regions of genomic DNA called enhancers that encode binding sites for transcription factor proteins. Binding of activators and repressors increase and reduce gene expression levels, respectively, but it is not understood how combinations of activators and repressors generate precise patterns of transcription during development. Towards this aim, we have developed high-throughput methods to dissect transcriptional enhancers to understand their role in determining patterns of gene expression in the fruit fly, a classic model system. We explore the role of enhancer architecture on gene expression by creating synthetic networks for pattern formation. Our work takes advantage of new genome engineering technologies, synthetic biology, high-throughput robotics, and cutting-edge single molecule imaging techniques in live embryos, creating a cross-cutting approach to developmental systems biology.
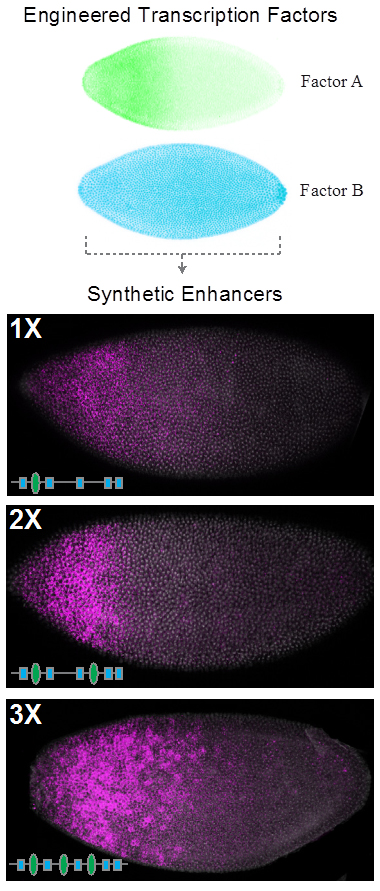
Enhancers are fundamental to development and evolution, yet we lack a detailed understanding of their necessary components, and how these components function collectively. We aim to determine the rules for the design and control of synthetic enhancers—including the number, affinity, and positioning of binding sites for transcription factors. We will explore this problem using a fully synthetic transcriptional platform in fruit flies consisting of engineered transcription factor gradients and artificial enhancers.
We aim to combine quantitative models of enhancer function with manipulations using engineered transcription factors to examine the extent to which enhancer function can be controlled in a predictable manner. We aim to develop models that allow the predictable control of enhancers, providing a framework for the quantitative control of enhancers with engineered transcription factors.
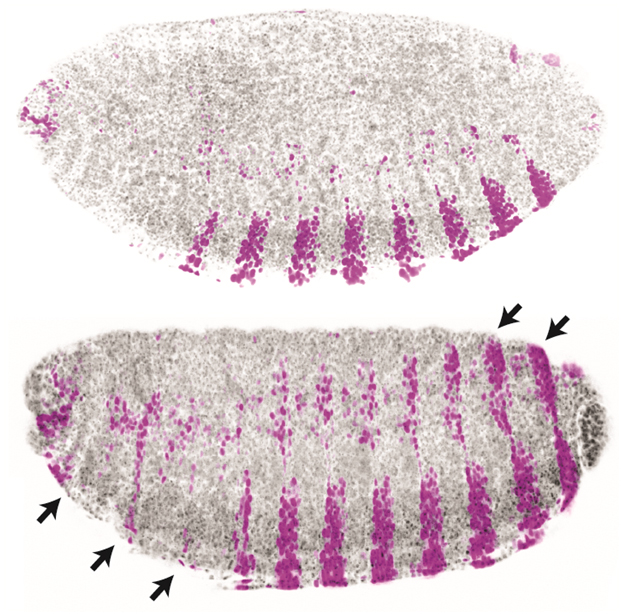
Hox gene regulation has presented a long-standing paradox because all Hox proteins bind to similar DNA sequences. We have discovered that Hox proteins bind primarily to clusters of low-affinity sites to regulate enhancers (Crocker et al., Cell 2015). Using a combination of classical genetics, live imaging, and super-resolution microscopy we aim to understand how these low-affinity sites are regulated in vivo.
Transcriptional networks contain the information required to confer robust positional information within a developing embryo. However, these networks have remained recalcitrant to modelling, as every parameter, such as diffusion and binding coefficients, are usually fitted values, and not derived from in vivo data. Building fully synthetic networks allows the exploration of network architecture unobscured by unknown elements. We aim to explore the role of network architecture on gene expression by creating synthetic networks for pattern formation.