
Moritz Gerstung
Visiting Group Leader
Birney Group
mg14 [at] ebi.ac.uk
ORCID: 0000-0001-6709-963X
EditMathematical analysis is crucial to the data-rich science of biology
Biology has become a data rich science. The amount and complexity of data produced in biology now exceeds that of any other scientific field.
As a consequence, statistical analysis and mathematical modeling are becoming ever more crucial ingredients in many areas of biology, including genetics, cell biology, structural biology, development and evolution.
Many groups at EMBL include researchers with a focus on mathematics and statistics, and several groups specialise in these fields:
Computational cancer biology

We have developed statistical models for relating different layers of genomic, molecular and clinical data to extract the precise connections among variables to understand the connection of genotype and phenotype. Moreover we have been working on biostatistical models and informatics tools for predicting outcome based on comprehensive high-dimensional data sets.
Another area of our research are the evolutionary dynamics of cancer. The process of developing cancer is driven by mutation and selection; hence the language to quantify that process is that of evolutionary dynamics. Deep sequencing unmasks the clonal composition of a cancer, which sheds some light on its evolutionary history. Accurate detection of subclonal mutations and reconstruction of phylogenies requires, however, accurate bioinformatics tools that we are actively developing.

Visiting Group Leader
Birney Group
mg14 [at] ebi.ac.uk
ORCID: 0000-0001-6709-963X
EditSystematic Biology 2015
64:e1-e25 Europe PMC
Bioinformatics 2014
30:1198-1204 Europe PMC
Blood 2013
122:3616-27 Europe PMC
Evolutionary analysis of DNA and amino acid sequences

We develop and use mathematical probabilistic models that describe DNA sequence evolution, DNA sequencing, and storage of digital information in DNA. The main focus of the group has traditionally been the development of models that describe how DNA changes through time during the course of evolution.
Our aim is to improve inference of evolutionary histories (phylogenies), and ancient genomes (ancestral sequence reconstruction), as well as to improve our capabilities at detecting the footprint of natural selection from genomic data.
More recently, the group has expanded its focus over computational and mathematical methods to improve the storage of digital information in DNA ─ a technology that promises to revolutionize how we store data in the long term. We are also developing probabilistic and information theoretical models to improve the efficiency of DNA sequencing ─ in particular nanopore sequencing.

Group Leader
Goldman Group
goldman [at] ebi.ac.uk
ORCID: 0000-0001-8486-2211
EditMolecular Biology and Evolution 2019
36, 2086-2103
Systematic Biology 2017
66, 769-785
Systematic Biology 2016
65, 328-333
Statistical Computing and Mathematical Modeling
Progress in biology is driven by technology. High throughput sequencing and microscopy require sophisticated statistical and computational operations in order to exploit their potential. To understand (and, eventually, manipulate) biological systems, all available data about them need to be integrated into computable maps and mathematical models. Ideas and techniques from physics, mathematics, statistics, computer science and engineering are the crucial drivers for our research.
Modern Statistics for Modern Biology

Group Leader and Senior Scientist
Huber Group
wolfgang.huber [at] embl.org
ORCID: 0000-0002-0474-2218
Edit2017
arXivBiostatistics 2019
DOI2019
Cambridge University PressComputational and evolutionary genomics
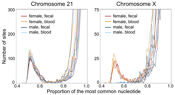
High-throughput sequencing is allowing the genome, transcriptome and epigenome of an enormous range of species, including model and non-model organisms, to be studied in exquisite detail.
Moreover, as technology develops further, we will move from studying populations of cells to studying regulatory processes at the single-cell level ─ this will enable numerous insights into developmental processes (e.g. embryogenesis and early-development), neurological processes (e.g., a fine-grained map of gene expression within specific brain regions), and the way in which tumours develop.
However, to make the most of these opportunities, appropriate computational tools for managing, analyzing, visualizing and downloading the data are essential. With this in mind, our work focuses on the development of statistical methods that will exploit these data to the fullest extent.
Genome Res. 2008
Sep; 18(9):1509-17 PDF
Nature 2010
Apr; 464(7289):768-72 PDF
Molecular Ecology 2010
(in press) (* joint first authors)
Statistical genomics and systems genetics
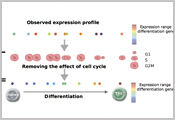
Our interest lies in computational approaches to unravel the genotype– phenotype map on a genome-wide scale. How do genetic background and environment jointly shape phenotypic traits or causes diseases? How are genetic and external factors integrated at different molecular layers, and how variable are these molecular readouts between individual cells?
We use statistics as our main tool to answer these questions. To make accurate inferences from high-dimensional ‘omics datasets, it is essential to account for biological and technical noise and to propagate evidence strength between different steps in the analysis. To address these needs, we develop statistical analysis methods in the areas of gene regulation, genome wide association studies (GWAS) and causal reasoning in molecular systems.
Our methodological work ties in with experimental collaborations and we are actively developing methods to fully exploit large-scale datasets that are obtained using the most recent technologies. In doing so, we derive computational methods to dissect phenotypic variability at the level of the transcriptome and the proteome and we derive new tools for single-cell biology.

Acting Head of AI centre
Stegle Group
ORCID: 0000-0002-8818-7193
EditNat Biotech 2015
33, 155-160 DOI
Nat Comm 2014
5, 5890 DOI
Nat Meth 2014
11, 817-820 DOI
From microscopy to mycology, from development to disease modelling, EMBL researchers cover a wide range of topics in the biological sciences.