BioSAXS at European Synchrotron Radiation Facility - Extremely Brilliant Source: BM29 with an upgraded source, detector, robot, sample environment, data collection and analysis software
J. Synchrotron Radiat. 30, 258-266 2023
Biological small angle X-ray scattering (bioSAXS) is a technique for obtaining ‘low’ resolution (~nm) structural information from macromolecules in solution that is complementary to Macromolecular Crystallography (MX) and becoming more widely used in structural biology. The bioSAXS beamline on BM29 at the ESRF has an optimised optical layout for solution scattering experiments with a multilayer monochromator for maximising the available flux and a toroidal focusing mirror for the generation of a small 200 x 100 µm2 (H x V) beamsize.
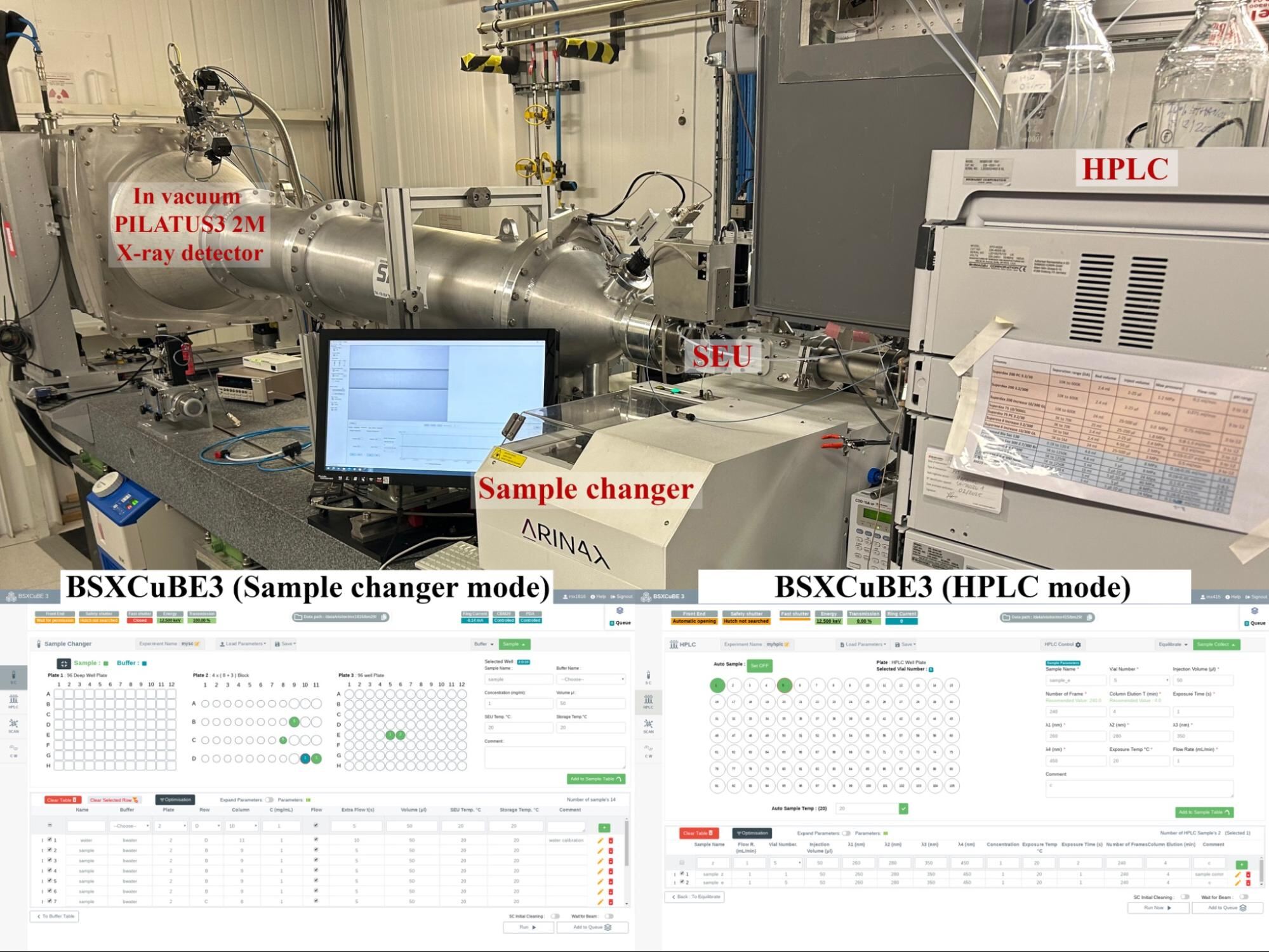
BM29 was recently upgraded to maximise its scientific potential after the ESRF- EBS upgrade and resumed full user operation in September 2020. BM29 is currently equipped with a new sample exposure unit (SEU) and liquid handling robot that were designed by the Instrumentation team as part of a trilateral agreement between the ESRF, EMBL Grenoble and EMBL Hamburg, as well as a newly installed in vacuum PILATUS3 2M X-ray detector. A Shimatzu HPLC system with an auto-sampler is also available for optimal experimental SEC-SAXS measurements from difficult biological systems. As part of the upgrade, an EMBL Full Stack Software Engineer, Jean Baptise Florial, from the Synchrotron crystallography team, was heavily involved in the development and implementation of BSXCuBE, a new web based graphical user interface (GUI) for BioSAXS experiments.
Currently, users can investigate a wide range of proteins under physiological conditions and undertake functional studies investigating the effects of differing temperatures (2-50 degrees ºC), pH (4-10), salt concentration and/or the addition of ligands in samples as small as 5 µl (25-30 µl are recommended). The bioSAXS beamline is dedicated to measuring biological macromolecules in solution with the aim of providing a simple to use facility with rapid access using the ESRF MX BAG or rolling application procedures.
A high level of automation is implemented to aid users with data collection, rapid preliminary analysis and online quality control using an open source EMBL-ESRF implementation of data analysis tools that are displayed in the Extended ISPyB for bioSAXS (Exi-ISPyBB) experimental database. This provides users with valuable feedback regarding their experiment to maximise throughput, dataset completeness and the possible need for additional measurements.
A second sample exposure unit is available to facilitate the measurement of SAXS data from microfluidic devices. Both static and mixing microfluidic devices can be accommodated, and both experimental types are currently being further developed and commissioned for user measurements. We have also recently started investigations into the development of an X-ray radical footprinting service to complement bioSAXS experiments. Both of these services are currently co-funded through the ARISE programme.
As part of the Partnership for Structural Biology (PSB) Small Angle Scattering (SAS) platform there is a unified application procedure within the ILL application procedure to enable joint SAXS/SANS experiments at the ILL and the ESRF facilities in one visit to the EPN site.
The BioSAXS beamline is run as a collaboration between the EMBL Grenoble Synchrotron Crystallography Team and the ESRF Structural Biology Group. See the beamline webpage for more information on the technical details and the ESRF website for beamtime application details.
J. Synchrotron Radiat. 30, 258-266 2023
J. Synchrotron Radiat. 29, 1318-1328 2022
17th Int. Conf. on Accelerator and Large Experimental Physics Control Systems (ICALEPCS'19), New York, NY, USA, WEPHA115 2019
In collaboration with the Structural Biology group at ESRF, we provide access to five macromolecular crystallography beamlines and one biological small angle X-ray scattering beamline at the ESRF.