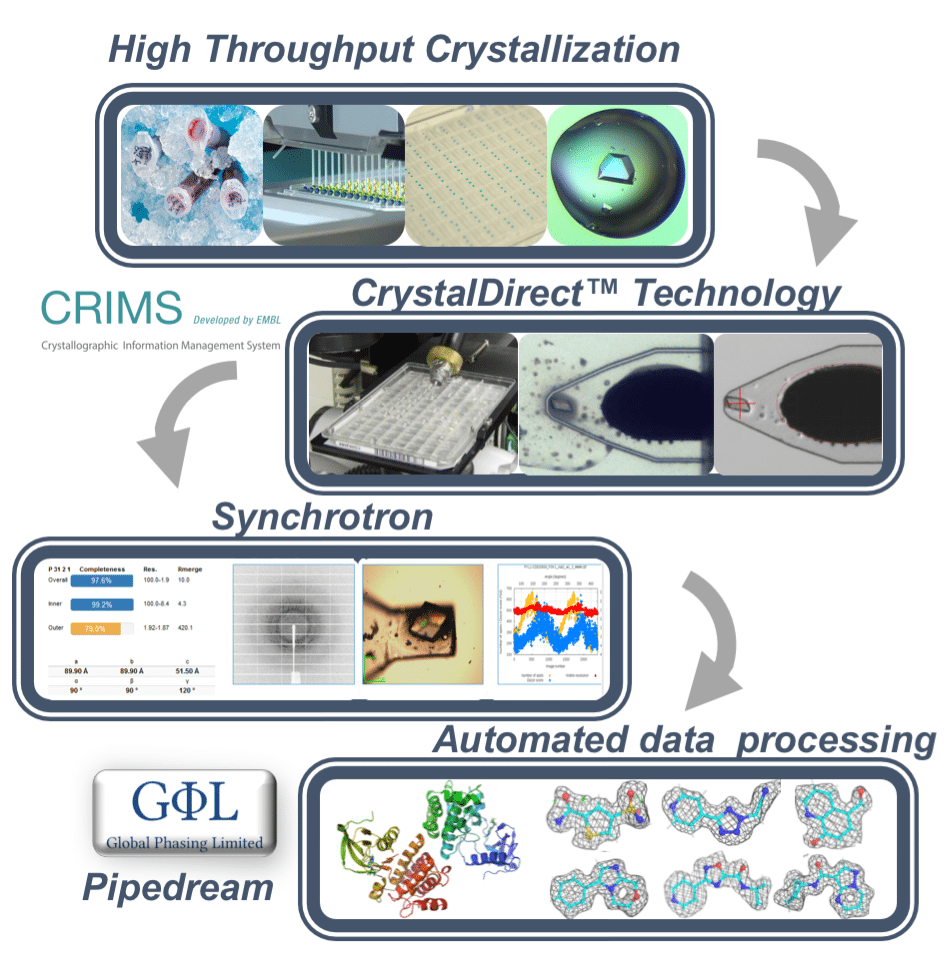
EMBL Grenoble operates the High Throughput Crystallization Facility (HTX lab), a large scale user facility offering High Throughput (HT) crystallography services. Since the start of operations in 2003 the HTX lab has provided services to over 800 scientists and processes more than 1000 samples per year.
The HTX lab has a strong focus in the development of new methods in macromolecular crystallography, including methods for sample evaluation and quality control and the creation of the CrystalDirect technology, enabling fully automated crystal mounting and processing.
The HTX lab has also developed the Crystallization Information Management System (CRIMS), a web-based laboratory information system that provides automated communication between crystallization and synchrotron data collection facilities, enabling uninterrupted information flow over the whole sample cycle from pure protein to diffraction data. This software is currently in operation in multiple laboratories across Europe
Through the combination of the CrystalDirect technology and the CRIMS software, the HTX lab has developed the concept of Online Crystallography: fully automated, remote controlled crystallography pipelines integrating crystallization screening, crystal optimization, crystal mounting and cryo-cooling, and automated X-ray data collection (at ESRF, PETRA III and SLS synchrotrons) into continuous workflows controlled though dedicated web interfaces. This approach minimizes the delay between crystal growth and measurement, accelerating the progression of challenging projects. The CrystalDirect technology also enables automated crystal soaking making it possible to support highly efficient automated, large-scale small molecule and Fragment Screening by crystallography.