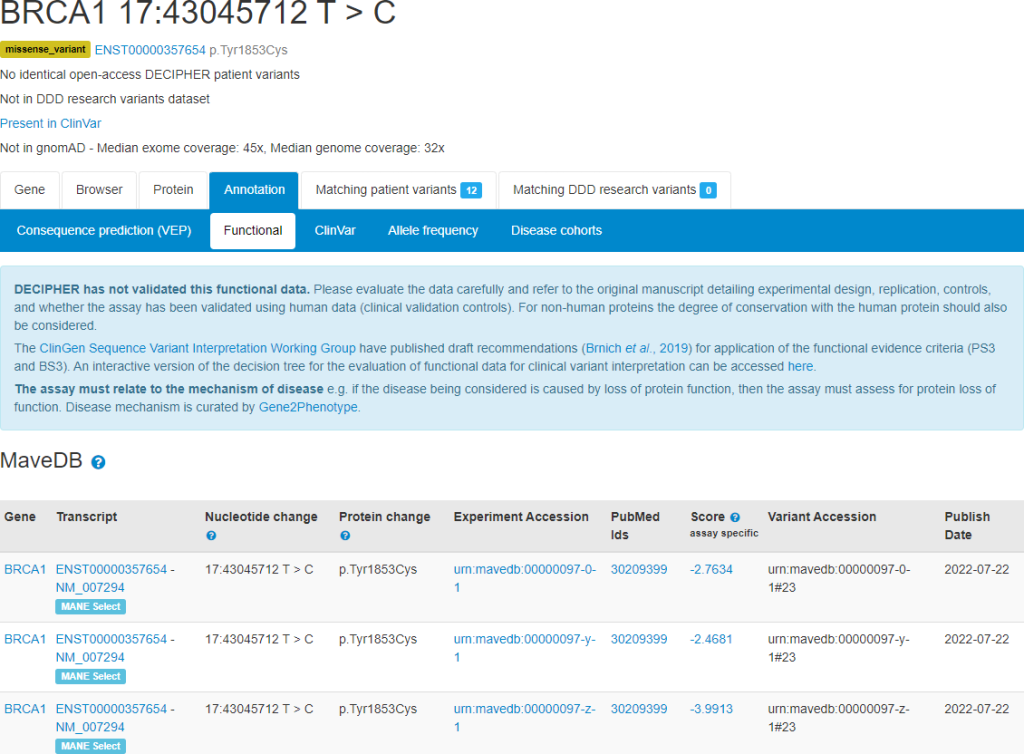
Multiplexed Assays of Variant Effect (MAVEs)
Functional data from Multiplexed Assays of Variant Effect (MAVEs), such as those generated by deep mutational scanning, from the MaveDB repository are now displayed on functional tabs. Links to experimental design and score sets in MaveDB are provided, in addition to publication links. This data can provide functional evidence for variant interpretation, in accordance with recommendations – a link to an interactive version of the decision tree for evaluation of functional data for clinical variant interpretation is available from this interface. DECIPHER makes use of reference sequence mapping and files created by Ensembl to display this data.
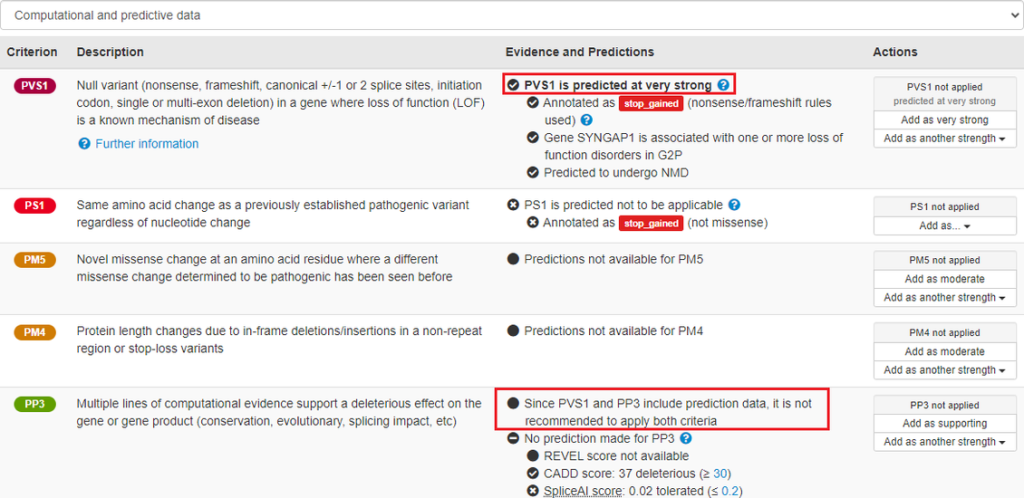
Sequence variant pathogenicity interface
In January DECIPHER released a new sequence variant ACMG/AMP pathogenicity interface which provides predictions for the application of criteria based on published guidelines. To ensure that predictions to apply a criterion are easily visible, this information is now displayed in bold. Text has also been added that states it is not recommended to apply both PVS1 and PP3. The user must use their expert knowledge of the variant data, and the patient, to decide if a prediction is valid for a criterion, and if relevant, accept it.
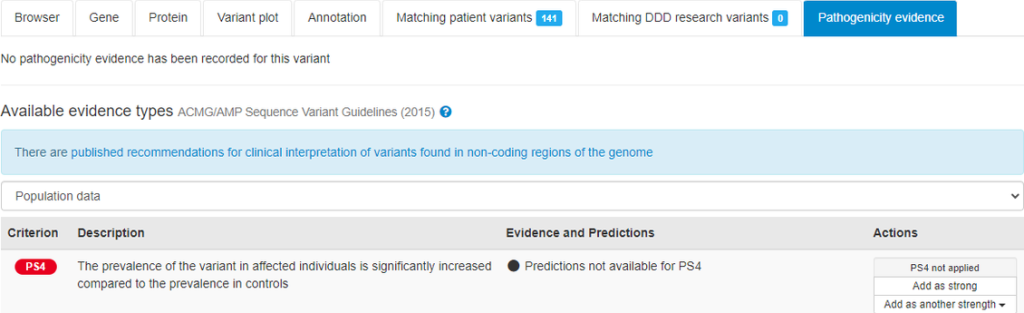
A link to the recommendations for clinical interpretation of variants found in non-coding regions of the genome is displayed in the sequence variant pathogenicity interface when a variant in a non-coding region is being evaluated.
The ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2024 were released in February 2024, and a link to these guidelines are now available from the sequence variant pathogenicity interface.
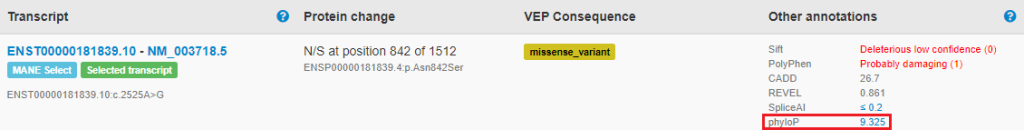
PhyloP scores
PhyloP scores which measure evolutionary conservation, are now displayed on consequence prediction tabs to assist variant interpretation. Conservation is important to consider when applying ACMG/AMP criterion BP7 for synonymous (silent) variants.
Gene names
The gene name has now been added to the protein browser to assist in easily identifying which protein is being viewed.
RefSeq transcript accessions
Refseq transcript accessions are now displayed in patient genotype tables, in the genome browser transcripts track and on the protein browser. Refseq transcripts are only displayed in these locations for transcripts in the MANE set, since these transcripts are identical to the corresponding Ensembl transcript.