EMBL’s European Bioinformatics Institute (EMBL-EBI) and Google DeepMind proudly present a significant update to the AlphaFold Protein Structure Database (AlphaFold DB) with new functionalities to make the data more discoverable.
AlphaFold DB has expanded to host over 200 million predicted 3D protein structures through Google DeepMind’s AlphaFold 2 system. The database has since undergone refinements to enhance user-friendliness and robustness to better support the scientific community. In a significant functionality update, two new data accessibility features are now available: sequence similarity-based search, and displaying structurally similar predictions on every AlphaFold DB prediction page.
These new functionalities of AlphaFold DB are available at https://alphafold.ebi.ac.uk/.
Sequence similarity-based search
In response to the requests from the user community since the initial launch of AlphaFold DB in July 2021, sequence similarity-based search functionality has now been introduced. Implementing this new feature, powered by the Basic Local Alignment Search Tool (BLAST), demonstrates the team’s commitment to enriching user experience and facilitating structural biology research.
The team has integrated BLAST into the search infrastructure to combine sequence similarity search results with existing search filters. With this update, AlphaFold DB search enables users to efficiently discover relevant predicted structures based on user-provided input protein sequences. The sequence search results have been integrated into the existing search results pages for intuitive navigation. Combined with the already available search filters, such as sequence review status and species, users can effectively identify proteins of interest.
Navigating the structural landscape with cluster members
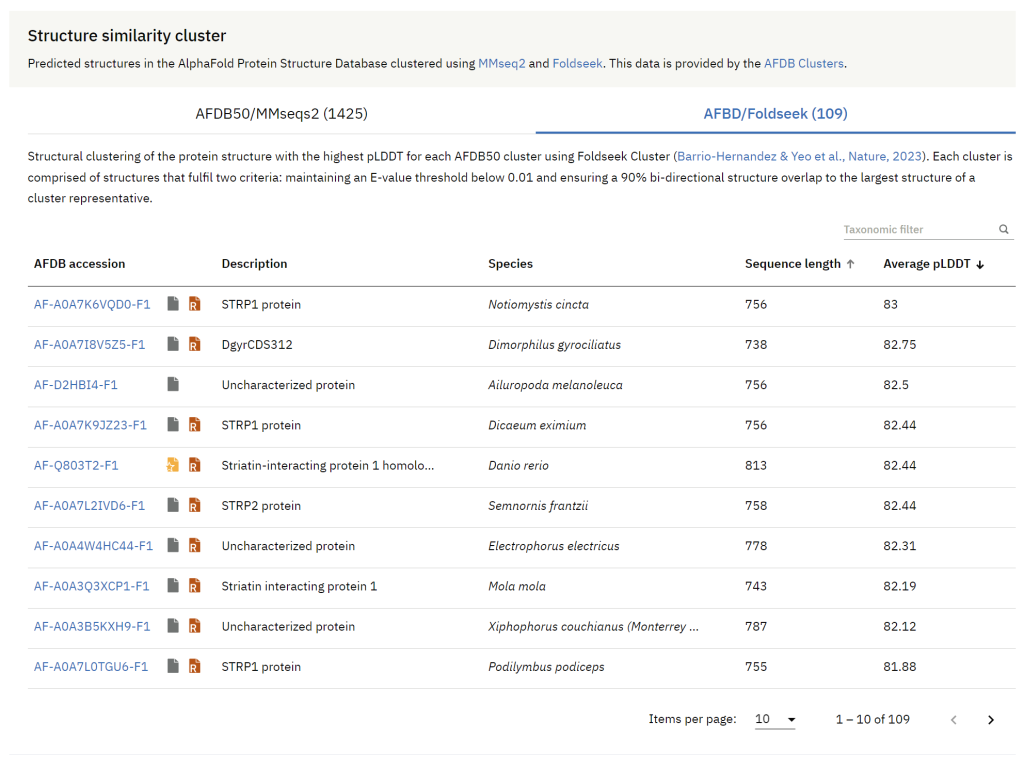
Acknowledging the challenges posed by hundreds of millions of predicted protein structures, the AlphaFold DB team collaborated with the Steinegger lab, to integrate Foldseek Cluster, an advanced structural alignment-based clustering algorithm for handling extensive datasets. These clusters are now available through a new structure similarity cluster feature, bringing similar structures directly to the prediction pages and ensuring a streamlined experience for researchers to explore the evolutionary breadth of proteins in structure space.
The clustering is a two step process. First, the 214 million UniProtKB protein sequences from the AlphaFold Database are clustered by MMseqs2, using a maximum of 50% sequence identity and a minimum of 90% sequence overlap. Representatives, chosen for their highest average pLDDT score, go through further clustering via Foldseek. These clusters from the second phase are filtered using an E-value cutoff of 0.01 and a minimum structural alignment overlap of 90%. This clustering approach aids users in identifying relationships through both sequence homology and structural resemblance across species.
Users now have direct access to the data generated in the two steps during the clustering process (AFDB50/MMseqs2 and AFDB/Foldseek) on the prediction page. Users can filter the data by species and sort the information based on sequence length and average pLDDT scores. We also indicate the sequence review status of a predicted structure (reviewed/unreviewed) and whether the protein is part of a reference proteome in UniProt.
For a more in-depth understanding of the clustering methods employed, please refer to the original publication, which provides detailed insights into the methodologies and processes applied.
Let us know what you think
The AlphaFold DB team is committed to continuously improving and meeting the needs of the scientific community. Let them know what you think of the new features, and provide suggestions via email at afdbhelp@ebi.ac.uk.
Edit