We are pleased to announce the release of Ensembl 110, and the corresponding release of Ensembl Genomes 57. This release brings exciting updates, such as the addition of regulation data to five animal genomes studied extensively in agriculture, the re-annotation of genomes in Ensembl Bacteria, and changes to REST API endpoints in our comparative genomics data. We have updated genomes across the different Ensembl sites, the addition of 15 rice varieties and invertebrate metazoan genome assemblies.
Can’t find a species you are looking for? Don’t forget that new genome assemblies and annotations are continuously added to Ensembl Rapid Release.
Vertebrates
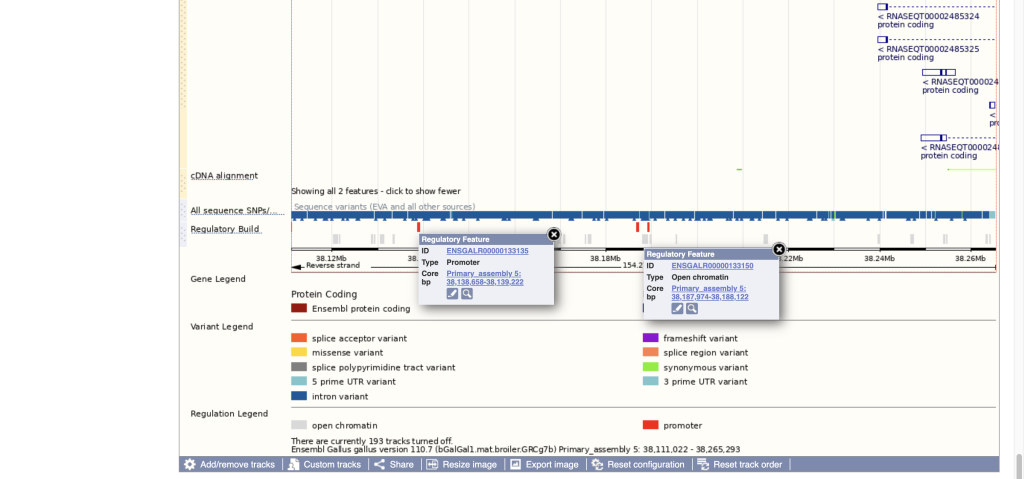
A major update in Ensembl is the addition of new regulation data. We have collaborated with the GENE-SWitCH and AQUA-FAANG consortia to add regulatory annotation in Pig, Chicken, Atlantic salmon, Turbot and European seabass. You can now visualise and find open chromatin regions and promoters in these species in the genome browser.
Three new plugins for the Ensembl Variant Effect Predictor (VEP) are now available:
- Geno2MP plugin highlights variant genotypes which have phenotypic profiles in the Geno2MP database. This will be available on the web interface, via the REST API and on the command-line.
- TranscriptAnnotator allows the annotation of variant-transcript pairs with custom data. This will only be available for command-line VEP.
- MaveDB plugin integrates experimentally determined measures of a variant’s effect from MaveDB. This will be available on the web interface, via the REST API and on the command-line.
We have also extended the analysis options available for structural variants (SV) in Ensembl VEP including more detailed molecular consequence predictions, more efficient integration of information from reference SV sets and support for breakend variant annotation and the integration of CADD-SV scores.
A release wouldn’t be complete without updates in human. We’re excited to tell you that the human genome assembly has been updated to the latest patch release GRCh38.p14. Note, however, that genes on patches will only appear on scaffold coordinates. Further, in the GFF3 annotation files, you will now find that MANE and Ensembl canonical attributes have been added as tags. Y pseudoautosomal region (PAR) genes are now stand-alone genes and are no longer taken from X, but MANE attributes remain on X PAR genes only.
Updated genomes
- Cricetulus griseus PICR: GCA_003668045.2 (Chinese hamster)
- Heterocephalus glaber: GCA_944319725.1 (Naked mole-rat, male and GCA_944319715.1 (Naked mole-rat, female)
New Rattus norvegicus (Norway rat) strains
- SHR/Utx RGD_8142385 GCA_023515785.1
- WKY/Bbb RGD_1581635 GCA_023515805.1
- SHRSP/BbbUtx GCA_021556685.1
Bacteria
This release brings an extensive update to Ensembl Bacteria. We introduce, for the first time, in-house gene annotation across bacterial genomes, through a collaboration between the Microbiome Informatics and Ensembl microbial groups at the EBI. Consistent annotation allows for better comparisons of prokaryotic species and pangenomes, and closer harmonisation with MGnify MAG (metagenomic assembled genomes) catalogues. Furthermore, the robust set of pipelines developed in this process allow Ensembl to address outdated and unannotated data sets in the prokaryotic space easily. You can read more about this in an upcoming Ensembl blog. Additionally, we used this chance to put Global Alliance for Genomics and Health (GA4GH) guidelines for systematic gene naming in place for the bacterial genes.
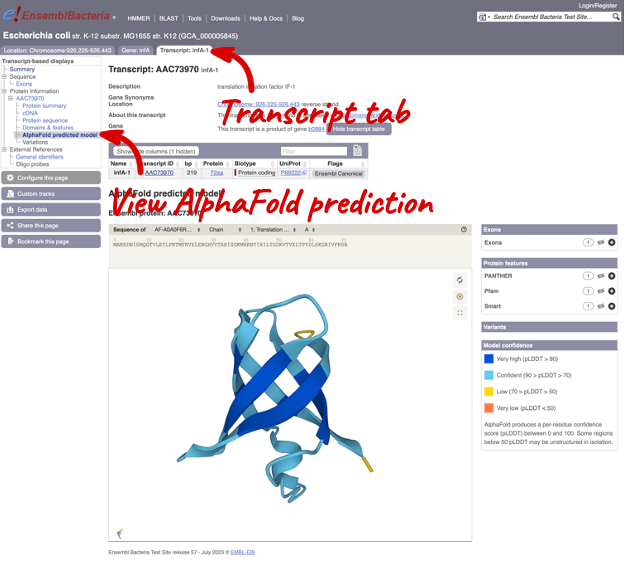
We are transitioning gradually to the new annotation in Ensembl Bacteria. We have updated annotations for all species in Ensembl Bacteria with the exception of 115 genomes, which represent widely cited community annotations or are model organisms. These 115 species, which are part of Ensembl’s pan-taxonomic comparative study, will not change for the foreseeable future, but now include AlphaFold predictions for proteins.
Plants
The new release includes a number of interesting additions to Ensembl Plants. For all wheat enthusiasts, we have added the Triticum aestivum IWGSC RefSeq v2.1, as well as the T. aestivum cv. Renan (GCA_937894285.1) assemblies to our database. You can find both genomes in the T. aestivum List of Cultivars. We have also updated Eragrostis tef (Teff) GCA_024500355.1 and Populus trichocarpa (Black cottonwood) GCA_000002775.4.
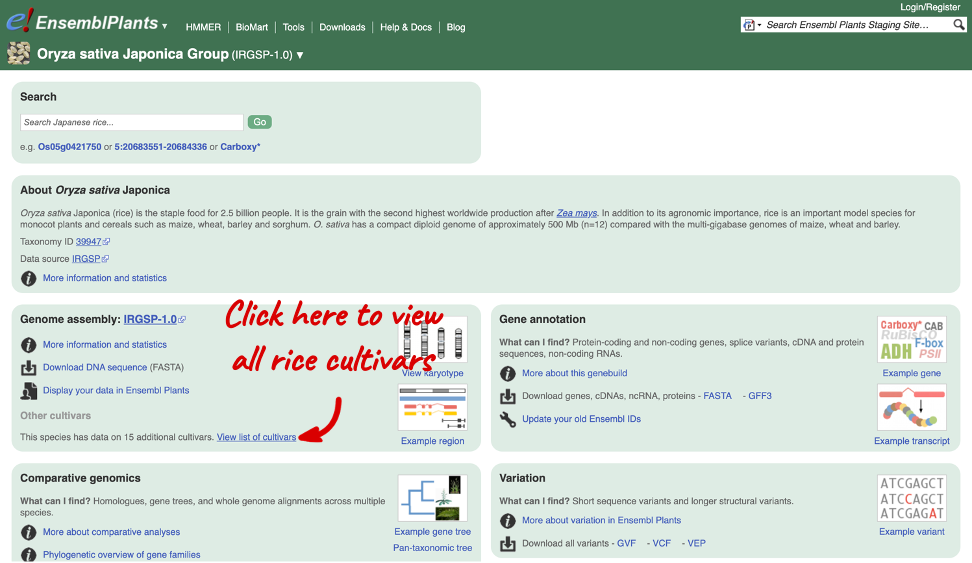
For rice, we have updated to the latest Oryza sativa (Rice) GCA_001433935.1 gene set and have added a whopping 15 cultivars! You can find all new cultivars in the O. sativa List of Cultivars (see screenshot below). These include:
- ARC GCA_009831255.1
- Azucena GCA_009830595.1
- Chao Meo GCA_009831315.1)
- Gobol Sail (Balam) GCA_009831025.1
- IR64 GCA_009914875.1
- Ketan Nangka GCA_009831275.1
- Khao Yai Guang GCA_009831295.1
- Larha Mugad GCA_009831355.1
- Lima GCA_009831355.1
- Liu Xu GCA_009829375.1
- MH63 GCA_001618785.1
- N22 GCA_001952365.2
- Natel Boro GCA_009831335.1
- PR106 GCA_009831045.1
- ZS97 GCA_001618795.1
Metazoa
Gene trees within Ensembl Metazoa have been expanded to cover 275 species by dividing them into 3 taxonomic clade sets: Metazoa, Protostomia, and Insecta. In addition, the release and update frequency of metazoan gene trees will change, with Metazoa and Protostomia being updated in every even-numbered release and Insecta being updated in every odd-numbered release. You can read more about this in an upcoming Ensembl blog.
You will also find updates in the following genomes:
- Amphimedon queenslandica (Sponge) GCA_000090795.2
- Acyrthosiphon pisum (Pea aphid) GCA_005508785.2
- Drosophila melanogaster (Common fruit fly) GCA_000001215.4
- Lingula anatina (Lamp shell) GCA_001039355.2
- Phlebotomus perniciosus (Sand fly) GCA_918844115.2
We have also added the following genome assemblies for existing species:
- Athalia rosae (Turnip sawfly) GCA_917208135.1
- Bombus terrestris (Buff-tailed bumblebee) GCA_910591885.2
- Diabrotica virgifera virgifera (Western corn rootworm) GCA_917563875.2
- Lucilia cuprina (Australian sheep blowfly) GCA_022045245.1
- Melitaea cinxia (Glanville fritillary) GCA_905220565.1
The invertebrate metazoa community have been busy releasing new genome assemblies, therefore, you can now find the following species in Ensembl:
- Amyelois transitella (Navel orangeworm) GCA_001186105.1
- Anthonomus grandis (Boll weevil) GCA_022605725.3
- Bicyclus anynana (Squinting bush brown) GCA_900239965.1
- Chelonus insularis (Parasitoid wasp) GCA_013357705.1
- Dermacentor andersoni (Rocky mountain wood tick) GCA_023375885.2
- Galleria mellonella (Greater wax moth) GCA_003640425.2
- Helicoverpa armigera (Cotton bollworm) GCA_023701775.1
- Helicoverpa zea (Corn earworm) GCA_022581195.1
- Homalodisca vitripennis (Glassy-winged sharpshooter) GCA_021130785.2
- Leguminivora glycinivorella (Moth) GCA_023078275.1
- Manduca sexta (Tobacco hornworm) GCA_014839805.1
- Neodiprion lecontei (Red-headed pine sawfly) GCA_021901455.1
- Neodiprion pinetum (White pine sawfly) GCA_021155775.1
- Nilaparvata lugens (Brown planthopper) GCA_014356525.1
- Pectinophora gossypiella (Pink bollworm) GCA_024362695.1
- Polistes canadensis (Red paper wasp) GCA_001313835.1
- Polistes dominula (European paper wasp) GCA_001465965.1
- Polistes fuscatus (Northern paper wasp) GCA_010416935.1
- Schistocerca americana (American bird grasshopper) GCA_021461395.2
- Sitophilus oryzae (Rice weevil) GCA_002938485.2
- Thrips palmi (Melon thrips) GCA_012932325.1
- Venturia canescens (Endoparasitoid wasp) GCA_019457755.1
- Zerene cesonia (Southern dogface) GCA_012273895.2
Other updates and highlights
- Compara REST API endpoints have been updated to require a species to be provided in addition to the stable ID. Read more about this change in this blog post.
- The Compara Perl API Compara::MemberAdaptor method fetch_by_stable_id has been removed and replaced by fetch_by_stable_id_GenomeDB, which accepts a Compara GenomeDB object in addition to a stable ID, and returns a corresponding gene or sequence member.
- The following options have been removed from the Regulation BioMart filters due to little use: karyotype band, marker and ENCODE pilot regions.
- Enhanced integration with AlphaFold protein structure database is now available: checksum-based mapping should ensure more stability and traceability of these external references. For technical reasons, mapping to protein structures is available for species with UniParc annotations only.
- Whole-genome alignments in Ensembl Fungi have been updated.
- Ensembl 92 archive has been retired.
This blog post was originally published on the Ensembl blog.
Edit