We have now expanded the structure superposition process on the PDBe-KB aggregated views of proteins to superpose the corresponding predicted model from the AlphaFold Protein Structure Database (AlphaFold DB). This new feature loads the model from AlphaFold DB on demand and superposes it onto the equivalent PDB structures for that UniProt accession, using the Mol* molecular viewer. This allows easy comparison of the experimental structures in the PDB with the predicted models in AlphaFold DB, to identify whether the predicted model matches a specific conformational state of the protein.
Experimental and predicted models superposed
You can view the structure superposition for a specific protein (per UniProt accession) at the PDBe-KB aggregated views of proteins. The superposition can be accessed from the Summary or Structures tabs using the green buttons labelled ‘3D view of superposed structures’. This opens the structure superposition window, with representative conformational states for that protein superimposed. The right hand menu gives an option ‘load AlphaFold structure’, which loads the AlphaFold DB model for this UniProt accession and displays it in the viewer.
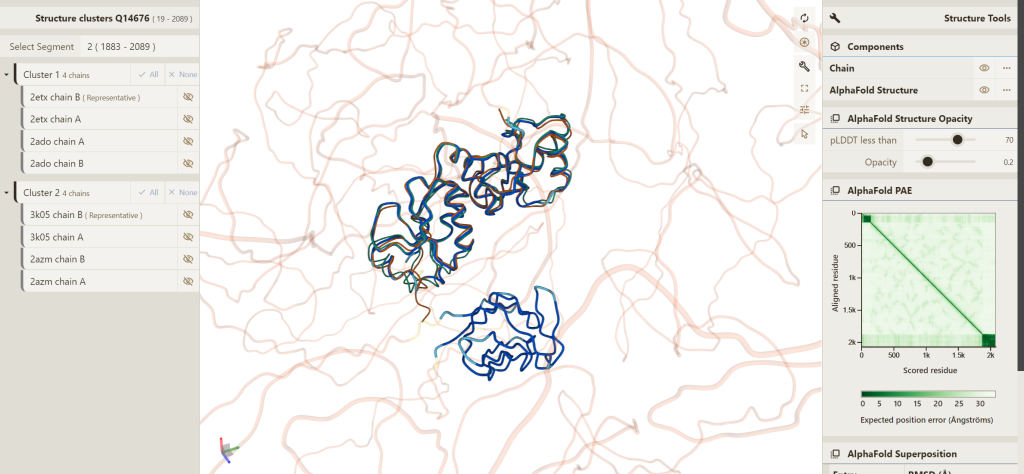
The AlphaFold model is displayed in ribbon format and coloured based on the pLDDT score, the per-residue confidence measure in the model prediction. In addition, the RMSD (Root mean square deviation) between the AlphaFold model and the best representative structure from each conformational state is shown, giving an indication of their similarity. The AlphaFold PAE plot is also displayed, which highlights the reliability of orientation of different parts of the structure relative to each other.
See full update on the PDBe website
Edit