Nassos Typas
Group Leader, Senior Scientist, Co-chair of Infection Biology and Microbial Ecosystems Transversal Themes
athanasios.typas [at] embl.de
ORCID: 0000-0002-0797-9018
EditSystems microbiology

Group Leader, Senior Scientist, Co-chair of Infection Biology and Microbial Ecosystems Transversal Themes
athanasios.typas [at] embl.de
ORCID: 0000-0002-0797-9018
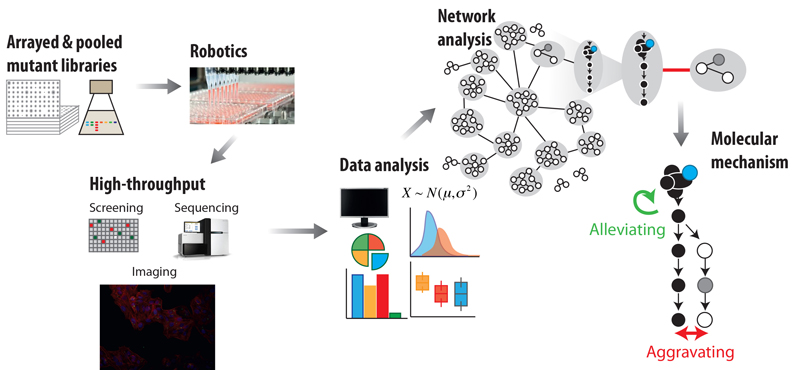
EditThe explosion of genomic sequence information provides a first step towards better understanding diverse bacteria, but also makes it crucial to develop large-scale approaches to characterise functions of novel genes and map them in pathways. We are developing high-throughput, multi-readout, automated approaches to quantitatively assess gene-gene, gene-drug and drug-drug interactions in many different bacteria and at many different levels (figure 1). We then use the data as starting points for generating mechanistic insights into targeted cellular processes, and also for uncovering how function, regulation and cross-talk between cellular processes changes across evolution, impacting the phenotype. Our main biological focus is on the bacterial envelope – its mode of assembly and growth, and its ability to sense the environment. Working at the intersection between genomics and molecular mechanism, we have discovered key missing players of major envelope pathways, uncovered niche-specific regulation of conserved envelope processes, detected how cell identifies malfunctions in such processes and identified linking proteins that allow different envelope machineries to coordinate their action.
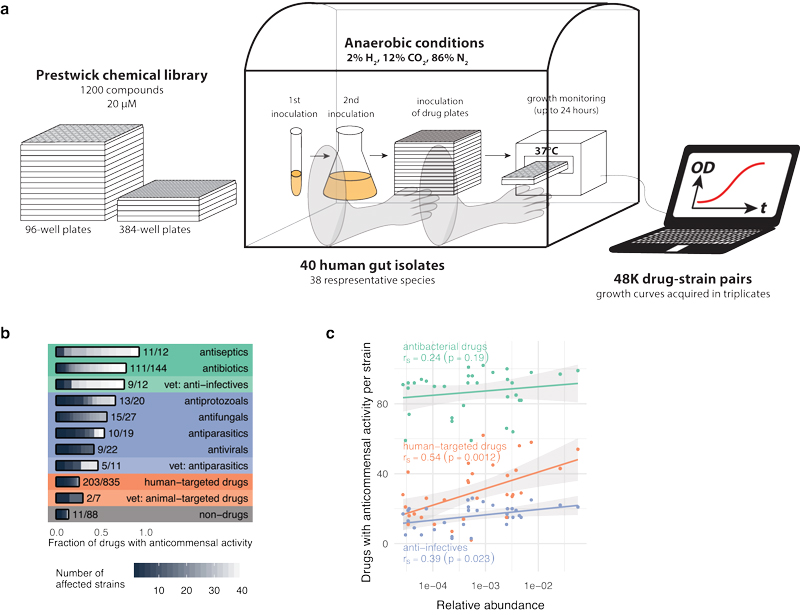
We have also recently moved our efforts to developing automated platforms and genome-wide approaches to study: (a) the mode-of-action and interactions of drugs, (b) the Salmonella-host interface, and (c) the human gut microbiome. Among other things, we have systematically profiled drug combinations in different bacterial species, unravelling general principles for these interactions, identifying adjuvants that re-sensitise multi-resistant pathogens, and dissecting the underlying mechanisms. In collaboration with the Bork and Patil groups, we developed one of the first comprehensive resources to study the gut microbiome, and used it to identify an unexpectedly high impact of non-antibiotic drugs on human gut microbes (figure 2). At least a quarter of such drugs directly inhibited the growth of gut microbes in vitro, causing antibiotic-like side effects in humans and potentially promoting antibiotic resistance.
We are broadly interested in the impact of mutations and horizontal gene transfer in the evolution of bacterial cellular networks. To tackle this, we are focusing on systems-based approaches in the E. coli pan-genome and in related enterobacteria. On the drug front, we are interested in discovering novel ways to target pathogens, in characterising the mode of action of new compounds, in predicting the outcome of combinatorial therapies and in understanding and mitigating antibiotic resistance. We are also continuing to work closely with different groups at EMBL (Bork, Patil, Savitski and Zeller) to develop integrated experimental and computational platforms for studying the gut microbiome. Here, we aim at understanding the assembly and dynamics of such communities, and the impact on them of drugs, natural and dietary compounds, physical parameters, and host molecules. Last, we are setting up a multi-pronged systematic approach aimed at gaining novel insights into the host-pathogen interface. We combine high-throughput reverse genetics, high-content microscopy and different types of quantitative proteomics to dissect the Salmonella-host interaction.